Jeffrey P. North and Dr. Ron Leavitt, Microbiology
In 1928 Alexander Fleming revolutionized the world with the discovery of penicillin. Penicillin and the antibiotics discovered thereafter revolutionized the treatment of infectious diseases. Equipped with these miracle drugs, scientists thought the battle against pathogenic bacteria was won. However, bacteria began to evolve to meet the antibiotic challenge. As the 20th century progressed, more and more bacteria developed resistance to one or more antibiotics. This resistance is achieved through mutation or through horizontal gene transfer, when one bacterium transfers the gene to another bacterium.
As a result of the antibiotic resistance phenomenon, the scientific community has began to explore alternative treatments. One possible avenue is to engineer nonpathogenic bacteria that can assume a role as resident flora and prevent colonization by invading pathogens. Such a strain could be administered as needed and would be self-perpetuating as it colonized the desired organism in a symbiotic relationship. However, extensive trials are necessary to ensure that the engineered strain is truly nonpathogenic and capable of withstanding challenge from various pathogens.
The addition of antibiotics to animal feed was implemented to stop food poisoning by preventing bacterial contamination. However, Salmonella poisoning from contaminated poultry products continues to be a problem throughout the world. In fact, many multi-drug resistant strains of Salmonella have begun to emerge. One strain, Salmonella enterica serotype typhimurium, has become resistant to chloramphenicol, streptomycin, ampicillin, sulfonamides, and tetracycline and has become a major enteric pathogen in both humans and animals (1).
Salmonella poisoning occurs when a bird is exposed to Salmonella bacteria which then multiply in the bird’s enteric tract. As the bird is processed into food products, the Salmonella contaminate the product and are ingested by an unknowing consumer resulting in food poisoning. Our goal is to stop this process at the initial step. We hypothesize that if we could colonize the birds’ intestines with a high-adhering, microcin-producing strain of nonpathogenic Escherichia coli, Salmonella would be unable to find a niche in the birds’ intestinal tracts.
We have isolated two wild strains of E. coli that we are using to engineer our poultry vaccine strain. The first strain produces a microcin that inhibits the growth of 58 of 59 strains of E. coli tested and 30 of 32 strains of Salmonella tested. The second strain exhibits a high-adhering phenotype to turkey intestinal epithelial cells. Both of the genes for these desired traits are located on plasmids, extrachromosomal circular DNA molecules. This paper focuses on the research dealing with the high adherence gene.
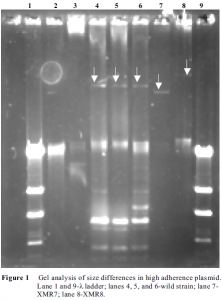
The high adherence phenotype is encoded on a rather large plasmid (approximately 108 kbp). To identify the gene or genes responsible for the high adherence phenotype, it was necessary to isolate the plasmid for analysis. First, the wild strain of E. coli exhibiting the high adherence phenotype was conjugated with a laboratory E. coli strain (XMR) that carries no plasmids. The resulting strains were screened using plasmid mini-prep analysis. Eight strains appeared to have received a plasmid approximately 108 kbp in size. These eight strains of XMR were then screened for a high adherence phenotype using green fluorescent protein (GFP) analysis on turkey intestinal epithelial cells.
Four of the eight strains exhibited high adherence when compared to the controls. Using these four strains, the plasmid was isolated in high concentration using the Qiagen Maxi Prep plasmid kit. Single and double digests of the plasmid were performed and used to generate a restriction map of the high adherence plasmid.
Two of the high adherence XMR strains were of particular interest. In GFP high adherence assays, XMR7 showed limited increase while XMR8 showed an extremely high degree of high adherence. Gel analysis shows that the plasmid in XMR7 is slightly smaller than the wild strain, but the plasmid in XMR8 is slightly larger (Figure 1). I believe conjugation with XMR7 terminated prematurely within a high adherence gene resulting in an incomplete high adherence phenotype.
Future research on this project will attempt to identify the exact location and character of the high adherence gene or genes through hybridization and cloning experiments. Further understanding of intestinal adherence will help not only in the engineering of a poultry vaccine strain of E. coli, but will also help researchers studying enteric pathogens like E. coli 0157H7.
References
- Glynn, M., Bopp, C., Dewitt, W., Dabney, P., Mokhtar, M., and Angulo, F. 1998. Emergence of Multidrug-Resistant Salmonella enterica Serotype Typhimurium DT104 Infections in the United States. N. Engl. J. Med. 338: 1333-1339.