Steven Alan Smith and Dr. Laura Bridgewater, Microbiology and Molecular Biology
Three chondrocyte-specific enhancer elements were previously identified in the mouse type XI collagen gene, Col11a2 (1-3). Enhancer elements are base pair sequences of DNA to which proteins bind enhancing expression of a particular gene. One of these enhancers, called F/G, is located in the first intron of that gene (3). This work has revealed the likely presence of additional positive cis-acting (enhancer) elements in the first intron, and one of those elements has been identified.
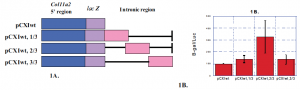
Fig. 1 The middle one-third of the Col11a2 first intron increased transcriptional activity (2). A) Portions of the Col11a2 first intron were cloned downstream of a lac Z reporter gene in the pCXI wt construct shown. A reporter gene produces an enzyme, in this case B-galactosidase(lac Z), that when mixed with substrate will produce light. The light produced measures the amount of expression of the gene. B) Transient transfection experiments, where above plasmids are put in rat chondrosarcoma, RCS, cells and light produced is recorded, were performed. Each experiment included co-transfection of a luciferase-expressing plasmid as an internal control for transf ection efficiency. Results are reported as B-gal/luc± S.E. The middle one-thrid, which contains the F/G enhancer, is most active.
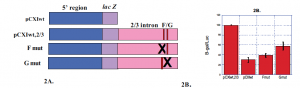
Fig. 2 The middle one-third of the intron contains other positive elements besides the F/G enhancer. A) Plasmid constructs designed for mutational and deletional analysis. B) Transient transfection in RCS cells, performed as in Fig. 1B. These experiments show that eliminating the entire intronic region decreases activity more than does the introduction of substitution mutations. The substitution mutations previously were shown to completely inactivate the enhancer.
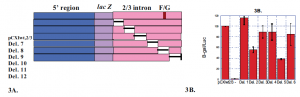
Fig. 3 The middle one-third of the Col11a2 first intron contains at least two positive enhancer elements. A) Plasmid contructs were made with 100-bp deletions in the middle one-third of the first intron as shown. B) Deletion constructs were tested in transcient transfection of RCS cells. Deletion 11, which eliminated the F/G enhancer, reduced activity as expected. Deletion 8 also reduced activity, suggesting the presence of a positive cis-acting element in the deleted region.
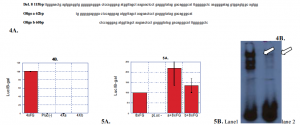
Fig. 4 Segments a and b contain no independent enhancer activity. A) Two smaller overlapping segments, a and b, of the Del. 8 region were synthesized and cloned into a luciferase reporter vector. B) The segments a and b were cloned as four tandem copies (multimerization) upstream of a minimal promoter and luciferase reporter in the p95Luc plasmid. A minimal promoter contains only the necessary sequences to which proteins bind and activate transcription, expression of the gene. These contructs were tested in transient transfection experiments in RCS cells. Neither one shows any independent enhancer activity. Four copies of the F/G enhancer clone into the same reporter plasmid served as a positive control, and the enhancerless plasmid, p95Luc, was the negative control. pSVB-galactosidase was co-transfected as an internal control for transfection efficiency, and results are reported as Luc/B-gal ±S.E.
Fig. 5 A 12-bp portion of the Del. 8 region interacts with the F/G enhancer to increase transcriptional activity. A) The segments of the Del. 8 region, a and b, were cloned upstream of the multimerized F/G enhancer in the 8xF/G luciferase reporter plasmid. 8xF/G alone served as positive control. In transient trasfections of RCS cells, the “a” element was shown to increase activity above that of the 8xF/G enhancer ~200% (More repeated experiments are needed to lower the standard error). Since the “a” element contains only 12bp that are not also present in the “b” element, those 12bp are hypothesized to bind proteins that interact with the F/G enhancer to increase transcription. B) Electro Mobility Shift Assays were performed to prove that proteins did bind to that 12bp region. The 12 bp are radiolabeled with a radioactive element, in this case p32. The radiolabeled contruct is then mixed with proteins extracted from the RCS cells. Competitor DNA is added to the mixtures to bind to non-specific proteins of the 12 bp’s. This mixture is then ran on an electrically charged gel seperating the different proteins that bind to the 12 bp. A band is seen where a protein binds to the radiolabeled contruct (lane 1). It can be proved that the protein is specific to the radiolabeled contruct by adding unlabeled 12 bp in a neighboring lane (lane 2) and comparing the bands. If the protein is specific the band will not be as strong in the lane with unlabeled 12bp (lane 2) because it is binding to both the unlabeled and labeled contructs.
In conclusion, athough the Del. 8 region of theCol11a2 first intron has no independent enhancer activity, it appears that a 12-bp sequence within that region interacts with F/G enhancer to increase transcriptional activity.
References
- Bridgewater, L.C., Lefebvre, V., and de Crombrugghe, B., Chondrocyte-specific enhancer elements in the Col11a2 gene resembel the Col2a1 tissue-specifice enhancer. J. biol Chem. 273, 14998 (1998).
- Bridgewater, L.C., Walker, M., Miller, G., Ellsion, T., Holsinger, D., Chen, R., Potter, J., Winkel, V., Sampson, J., Shang, Z., Mckinney, S., de Crombrugghe, B., (submitted 2001).
- Liu, Y., Li, H., Tanaka, K., Noriyuki, T., & Yamada, Y. (2000). Indentification of an enhancer sequence within the first intron required for cartilage-specific transcription of the a2(XI) collagen gene. Journal of Biologiacal Chemistry, 275, 12712-12718.