Aaron Madsen and Dr. Keith Crandall, Integrative Biology
Astacopsis gouldi has been known to weigh up to 6 kg, and live for as long as 60 years (Horwitz and Hamr, 1988). However, fishing pressure, as well as habitat destruction and pollution have had detrimental effects on A. gouldi populations. The populations of the species in accessible rivers have declined, as have the number of males reaching sexual maturity. These factors led to the placement of A. gouldi on the endangered species list in 1996 (IUCN, 1996). Conservationists are considering the relocation of crayfish from populations in more remote rivers and streams where the decline in numbers has been less severe, to streams where the population has been all but wiped out. Two major concerns with this process are whether or not transplanting animals from one population to another would disrupt the genetic structure and associated evolutionary processes, and whether or not non-adapted varieties would be relocated. A gouldi is found in independent river systems throughout northern Tasmania with the exception of the Tamar River system, which is located in the center of northern Tasmania (Horwitz, 1991). This has resulted in a split distribution of the species. The independence of the river systems and the split distribution has led us to investigate the following questions:
- What is the level of genetic diversity in the species?
- How much gene flow exists between the independent river drainage systems and between the systems separated by the Tamar River system?
- What is the genetic population structure of the independent river drainage systems, and the split distribution caused by the absence of animals in the Tamar River system?
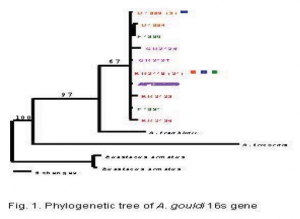
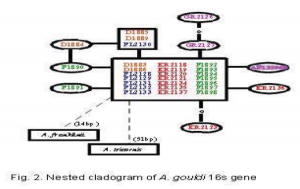
To answer these questions, a total of 32 animals from 6 locations across northern Tasmania were captured. Their DNA was extracted and sequenced for the 16s, COI, and COIII genes as was described in the proposal. The computer program ModelTest v3.0 (Posada and Crandall, 1998) was used to determine the model of evolution for the 16s gene region. We then used the computer program PAUP* 4.0 (Swofford, 2000) to estimate a phylogenetic tree, implementing the best fit model HKY+G. I used a cladogram to help resolve those parts of the phylogenetic tree which were unresolved. The COI and COIII data is nearly complete and will be added to the 16s data for publication.
At the present, we have 446bp fragments of the 16s gene from 32 individuals, 650bp fragments of the COI gene for 23 individuals, and 550bp fragments of the COIII gene for 17 individuals. We chose to analyze only the 16s data at this time, but we plan to include the data from the other two genes as soon as they are completed. Our results show that there is very little genetic diversity in Astacopsis gouldi. For the 16s gene, there were only 10 haplotypes from 32 individuals, differeing by a maximum 1.1% sequence divergence. One of those haplotypes represented 21 of the individuals. Our preliminary COI and COIII data also show about 1% sequence divergence, which is considered low for animals of the same species. All of the A. gouldi sequences formed a mono clade with respect to 2 congenerics (Fig. 1), and there is no genetic population structure among the haplotypes, although the 2 haplotypes from east of the Tamar River (GR2126 and GR2127) were not found west of the Tamar River (Fig. 2). From this 269 data, we are able to conclude that the 16s, COI, and COIII genes have low levels of variation in A. gouldi. We also conclude that there is high gene flow among different populations, and that relocation of A. gouldi for conservation purposes would do little to affect the genetic population structure of the species, since it is all but absent. There is still some work to be done before this project will be finished. We will shortly finish the COI and COIII genes, and we are considering using another gene to provide us with more data. I have learned much about evolutionary genetics and how they can be used in conservation efforts, as well as how computers can be used to interpret data.
References
- HILTON-TAYLOR, C. (compiler) 2000. 2000 IUCN Red List of Threatened Species. IUCN, Gland, Switzerland and Cambridge, UK.
- HORWITZ, P. 1991. On the distribution and exploitation of the Tasmanian giant freshwater lobster Astacopsis gouldi Clark. A Preport to the Office of the National Estate.
- HORWITZ, P. and P. Hamr 1988. Papers and Proceeding of The Royal Society of Tasmania 122:69-72.
- POSADA, D. and K.A. CRANDALL, 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14 (9): 817-818.
- SWOFFORD, D.L. 2002 PAUP* 4.0b10. Phylogenetic analysis using parsimony and other methods. Sinauer Assoc. Sunderland, MA.