Trever Bradley Burgon and Dr. Gregory F Burton, Microbiology
Human Immunodeficiency Virus (HIV), the retrovirus that causes Acquired Immunodeficiency Syndrome (AIDS), has infected over 60 million people and become what the Joint United Nations Program on HIV/AIDS calls: “the most devastating disease mankind has ever faced” (8). Current treatment, known as HAART (highly active antiretroviral therapy), is able to reduce HIV in the blood to undetectable levels but it cannot completely block replication nor remove HIV from the body. When patients stop taking HAART, their viral load (the quantity of virus detected in their blood) and symptoms quickly return to pretreatment levels. This viral rebound is caused by latent virus that persists in reservoirs that are untouched by the drugs. These reservoirs harbor and protect HIV throughout infection and reignite infection once HAART is terminated.
Unique cells in the body, called follicular dendritic cells (FDC), have the unusual ability to trap HIV on their cell surface where the virus is unaffected by HARRT. HIV particles can be seen covering the FDC of infected patients, and recent evidence shows that this HIV remains infectious for long periods of time (5). These data suggest that FDC are a long-term reservoir of infectious HIV.
In order to better understand the role of FDC as HIV reservoirs I used DNA sequence analysis to compare the genetic relationship of HIV isolated from FDC (F-HIV) with HIV isolated from productively infected CD4+ T cells (T-HIV) of an AIDS autopsy patient (patient 559). If FDC are an HIV reservoir, they should contain an HIV population that is genetically more diverse and evolutionarily more ancient than that of the CD4+ T cells.
In this study we sequenced a 1029 base pair sequence of the HIV gene pol. The section we sequenced includes regions encoding the viral enzyme PR (protease) and a portion of the viral enzyme RT (reverse transcriptase). Both of these viral enzymes are targets of HAART drugs, allowing us to use the sequence also track the evolution of specific drug resistant mutations.
Materials and Methods
HIV DNA isolated from coculture assays was amplified by nested-set PCR. The amplified products were then ligated into the plasmid vector PCR3.1 (Invitrogen). The cloned vector was transformed into STBL2 bacteria (Invitrogen) by heat shock and grown overnight on LB agar plates. Individual colonies were then isolated. Presence of the 1029 bp insert was verified by EcoRI digestion and agarose gel migration. The cloned plasmids were then isolated using DNA Mini-Prep (Qiagen) and sequenced by Big-dye chain termination chemistry (Applied Biosystems). Sequences were edited and verified using the computer program SEQUENCER (Gene Codes Corp., Ann Arbor, Mich). Nucleotide alignments were done with CLUSTAL W (7) and then edited and gap stripped with MacClade (2). The best-fit evolutionary model was selected in MODELTEST (4) and determined to be HKY+I+G. This model was then used in PAUP* (6) to construct a maximum likelihood (ML) tree.
Results and Discussion
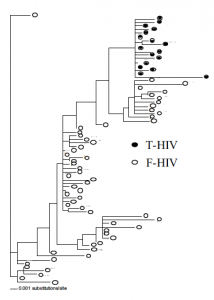
The maximum likelihood phylogenetic tree (Fig. 1) shows that F-HIV is not only clearly distinct from T-HIV isolated from the same tissue, but also appears to be more diverse and more ancient. These findings are consistent with the original hypothesis that FDC trap and preserve HIV for extended periods of time. T-HIV clones represent the most recently successful HIV because they come from cells that are currently infected and producing virus. FDC, however, have been trapping and preserving successful mutations throughout the infection. It appears that clones in the bottom left of the tree are the evolutionary ancestors of clones in the top right. This analysis supports the hypothesis that FDC trap and preserve infectious HIV in the human body.
Over a dozen different resistance mutations were found in the sequenced clones. The following are highlighted: an R57K mutation in PR confers resistance to the drug Nelfinavir (1). This resistance mutation does not occur in the T-HIV, but it is found in eight of the F-HIV clones. In the viral enzyme RT, a R211K mutation results in AZT resistance (3). While all the T-HIV clones are sensitive to AZT at this codon, nine F-HIV clones show resistance. These data are difficult to interpret at this time because we do not know the medical history of the patient or when the patient took the different HAART drugs. The resistance mutations found only in FHIV may represent mutations that arose when the patient underwent therapy and but then disappeared when the therapy changed or was ceased. Without that selective pressure of the drugs, the replicating virus would have mutated while the FDC would have trapped and preserved the older, resistant clones.
This study sheds new light on the role of FDC as HIV reservoirs. FDC-trapped HIV is phylogenetically more distinct and ancient that HIV found in T cells. In the future, I look forward to the publication of these data in a scientific journal. As we better understand how HIV acts in the body, we come closer to a cure for the disease.
FIG. 1. ML phylogenetic tree showing the evolutionary relationship of F-HIVand T-HIV clones. 17 T-HIV clones and 57 F-HIV clones are depicted.
References
- J. Lawrence, J. Schapiro, M. Winters, J. Montoya, A. Zolopa, R. Pesano, B. Efron, D. Winslow, T. C. Merigan. 1999. J Infect. Dis. 179, 1356 (1999).
- D. R. Maddison, W. R. Maddison. Sinauer Associates, Inc., Sunderland, Mass. (2001).
- V. Miller, A. Phillips, C. Rottmann, S. Staszewski, R. Pauwels, K. Hertogs, M. P. de Bethune, S. D. Kemp, S. Bloor, P. R. Harrigan, B. A. Larder. J. Infect. Dis. 177, 1521 (1998).
- D. Posada, K. A. Crandall. Bioinformatics. 14, 817 (1998).
- B. A. Smith, S. Gartner, Y. Liu, A. S. Perelson, N. I. Stilianakis, B. F. Keele, T. M. Kerkering, A. Ferreira-Gonzalez, A. K. Szakal, J. G. Tew, G. F. Burton. J. Immunol. 166, 690 (2001).
- D. L. Swofford. Sinauer Associates, Sunderland, Mass. (2001).
- J. D. Thompson, D. G. Higgins, T. J. Gibson. Nucleic Acids Res. 22, 4673 (1994).
- UNAIDS. 2002. AIDS epidemic update: December 2001. UNAIDS. [Online.] http://www.unaids.org/epidemic_ update/report_dec01/index.html#full.