Mo Lee and Dr. John Kauwe, Department of Biology
It turns out that the data for chromosome 21 didn’t come to me at all. There were some problem with the data itself according to our co-researchers. However, the data for the phenotype YKL40, which is a promising gene that might affect the pathology of Alzheimer’s Disease. Dr. Kauwe decided that we should utilize the ORCA grant on this data that we had.
Method
For both YKL_Plasma and YKL_CSF, I performed linear regression analysis with covariates 1) age, PC1All, PC2All, and 2) age, APOE, PC1All, PC2All. Four analyses are performed. I included APOE just in case we might need it later in the analyses. APOE after step-wise analysis is not significant enough to be included as covariate. YKL 40 is not normally distributed after normality test in SAS. So I log-transformed it once. There are 982617 makers in the dataset. After Bonferroni correction, the threshold should be 5.1*10^-8. Results are attached as excel spreadsheet and there is a discussion below the SAS stats data.
Four analyses as below:
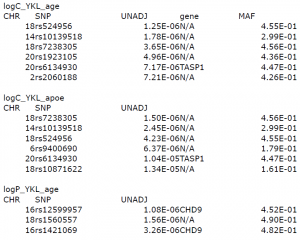
- logC_YKL_age: CSF samples with age, PC1All, PC2All covariates
- logC_YKL_apoe: CSF samples with age, PC1All, PC2All, APOE covariates
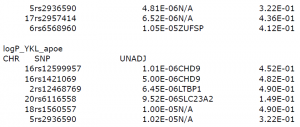
- logP_YKL_age: plasma samples with age, PC1All, PC2All covariates
- ogP_YKL_apoe: plasma samples with age, PC1All, PC2All, APOE covariates
Results
Discussion
For log_C_YKL samples, the three tophits SNPs are either on chr 18 or chr 14. (rs524956 and rs7238305 on chr 18, rs10139518 on chr 14) These SNPS are not in any gene coding region.
For log_P_YKL samples, rs12599957 and rs1421069 on chr 16 and rs1560557 on chr 18 are the three tophits. The SNPs on Chr 16 are in CHD9 gene region.
I think it will be worth a try to look closely at the regions where those SNPs locate on chr 14, 16, and 18. Either we can do set-base analyses on those regions or we can just increase sample size if possible because our P values are kind of close to the threshold. These data I produced can only tell us that they might be potential biomarkers for Alzheimer’s disease. Much needs to be done in the wet lab because it is only computational result. Also, we don’t have enough sample size to make the P value significant enough.