Maren Ettinger and Dr. Keith Crandall, Department of Biology
The purpose of phylogenetics is to understand the evolution of a group of organisms and to resolve the relationships within groups. Once phylogenies are created, they can be used for a number of different analyses. My project focused on using a phylogeny of the infraorder Anomura to create new insight into the evolution of the group. We examined deep phylogenetic relationships using molecular and morphological data, detected diversification shifts in the history of the anomurans, and traced character (habitat type, body shape) traits across the phylogeny. Anomurans represent a speciose group of decapod crustaceans. They include hermit crabs, yeti crabs, squat lobsters, king crabs, porcelain crabs, and hairy crabs. Because they are so diverse, the relationships within the anomurans are unresolved and misunderstood. The anomurans are economically important in aquaria and seafood trade (hermit crabs, king crabs).
The main goals of my project were to create a robust phylogeny from morphological and molecular data, to include fossils to date the tree, reconstruct ancestral states of the anomurans (i.e., body form and habitat type), and detect diversification shifts and relate them to morphological characters and geological time. This project has allowed me to work on an international level with the world’s leading researchers in the field of systematics. We were able to complete these goals and are in the advanced stages of submitting our manuscript to an academic journal.
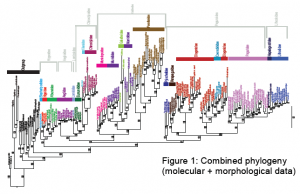
The Bayesian analysis from the combined (molecular + morphology) dataset recovers Anomura as a monophyletic group with high support (100 = Pp, Fig. 1). The majority of the nodes (86%) are recovered with very high support (>95). Three families are recovered as para- or polyphyletic (Diogenidae, Paguridae, Munididae). With the exception of three families (Blepharipodidae, Kiwaidae, Lomisidae- each having a single representative), the remaining families were found to be monophyletic with high support. We recovered the most robust anomuran phylogeny to date in both taxon sampling and gene depth. It was a unique experience for me, as an undergraduate, to participate and play an important role in such a large project and international collaboration.
The anomurans diverged from their closest ancestor (the Brachyuran crabs) approximately 259 million years ago (MYA). The most recent common ancestor (MRCA) of the anomurans dates at approximately 221 MYA. These dates are concordant with the fossil record of anomurans and brachyurans. We were able to combine molecular, morphological, fossil, and computer-simulated data to better understand how this group evolved and became so diverse.
Our ancestral reconstruction analyses recovered that a crab-like ancestor gave rise to all extant anomuran lineages. In addition to the earliest branching clade (Blepharipodidae, Hippidae, Albuneidae), carcinization (crab-like form) arose independently three times during the evolution of the group, twice through squat lobsterlike intermediaries and once through a symmetrical hermit crab-like ancestor. The transition into freshwater occurred once in the family Aeglidae and the transition into a semi-terrestrial habitat occurred once in the family Coenobitidae. Both transitions occurred through marine ancestors. This was an interesting result because there must be something advantageous about the crab-like form for it to have evolved three
separate times.
Diversification of the anomurans is fairly uniform. We detected two shifts in diversification-one in the family Lomisidae and one in the squat lobster family Chirostylidae. This suggests that body form affects the rates of speciation. This is an area that would require further research to make more definitive claims.
This project encountered very few problems along the way. The only problems that arose were from the computer analyses and mostly involved designing code correctly. This project has also taken longer to get submitted to a journal than we expected. This was simply because revising has taken a long time. We are currently in the late stages of revising and will soon submit a manuscript to the journal Cladistics. Cladistics is a high impact journal and we hope people will find our results interesting and exciting.
Working in a research lab, and on the anomuran project specifically has been an irreplaceable part of my education at BYU. Doing research in the Crandall lab is ultimately what pushed me toward pursuing a PhD in Human Genetics. Being in a research environment, where undergraduates are encouraged to pursue their own research is a unique opportunity and an area that BYU excels in. Learning to collaborate with researchers across the world has given me better insight into the scientific community and the importance of collaboration. Being part of the ORCA program has allowed me to have these experiences and gain the research experience I need to enter graduate school.