Alan Colver and Dr. Keith Crandall, Department of Biology
The taxonomic group Pancrustacea encompasses more than half of the world species. The two main groups within Pancrustacea are the crustaceans and hexapods (insects). Understanding these two groups will aid in preservation, population control, and economic pursuits involving these groups. In our evolutionary analysis, six different genes, which are good indicators of evolutionary divergence (Felsenstein, 2003), were analyzed to gain a much deeper understanding of the relationships among these species than ever before and to hopefully resolve much of the controversy in the scientific community concerning their evolution (Jenner, 2010).
The two major processes in our study were data collection followed by data analysis. An online repository known as GenBank already contained many sequences of our species of interest (dark blue bars in Table 1); however, many more samples were needed to perform both a broad and in-depth analysis; these are the samples that the Crandall lab collected on several expeditions (light blue bars in Table 1) and sequenced using a novel technique developed at BYU (Bybee, 2011).
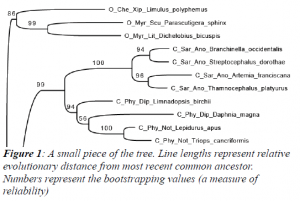
After collection the data were processed. This included aligning (matching up the DNA sequences) the data using MAFFT (Katoh, 2002) followed by a comparative analysis (on BYU’s super computer) of the differences between the sequences using a maximum likelihood statistical model known as RaXML (Shimodaira, 1999). The output of this final step results in a hypothesized evolutionary tree (a small excerpt can be seen in Figure 1). These steps were repeated several times, each time including more data and corrected mistakes in previous data.
The first trees had some major flaws, which we quickly found to be contaminated data or from an unreliable source. These were removed, and by the 4th iteration we had produced a fairly robust hypothesized evolutionary tree. This was then passed on to several collaborators, each being an expert in one or more orders (smaller sub-groups of Pancrustacea). With their feedback and additional data, the tree grew in comprehension and accuracy. To date, we are currently on our 7th iteration and likely only a few more iterations will be necessary prior to publication.
Despite not being ready for publication immediately, several conclusions can be drawn with the current analysis. The hotly disputed order Remipedia (Jenner, 2010) appears to be monophyletic (having all modern-day species derived from only one common ancestor) as opposed to paraphyletic (from multiple ancestor species) and the sister group of Hexapoda. This conclusion is still being analyzed for certainty before publication. Additionally, we can provide strong evidence for the true relationships among many other orders within Pancrustacea.
One of the weaknesses is of the study is the slight subjectivity in determining bad DNA sequences. Some sequences are very obviously contaminated in the fact that they match much closer to bacteria than any pancrustacean; however, there were some sequences that were extremely different from any pancrustacean and dissimilar from bacteria. This presented a challenge of determining their accuracy, and we left the decision for exclusion to each of the respective experts.
Given the vast amounts of data needed to be gathered and analyzed and given the computational expense of these analyses, this project is intrinsically lengthy. Despite this, we have made significant headway this last year and appear to be on track to publish in the latter half of 2013.
The ORCA Grant has enabled me to commit more time to this project than otherwise would have been possible.
References
- Bybee, S., H. Bracken-Grissom, B. Haynes, R. Hermansen, R. Byers, M. Clement, J. Udall, K. A. Crandall. 2011. Targeted amplicon sequencing (TAS): A scalable next-gen approach to multi-locus, multi-taxa phylogenetics. Genome Biology and Evolution 10.1093/gbe/evr106.
- Felsenstein, Joseph. Inferring Phylogenies. 2. Sinauer Associates, 2003. Print.
- Jenner, R. A. (2010). Higher-level crustacean phylogeny: Consensus and conflicting hypotheses. Arthropod Structure & Development, In Press, 1-11.
- Katoh, Misawa, Kuma, Miyata 2002 (Nucleic Acids Res. 30:3059-3066) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform.
- Shimodaira, H. and Hasegawa, M. (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference, Molecular Biology and Evolution, 16, 1114-1116