Joshua K. Sailsbery and Dr. David A. McClellan, Integrative Biology
Detecting selection on a molecular level has a wide range of applications. Knowing the selection that has occurred may lead to reconstruction of phylogeny trees, better understanding of the changes a given protein is experiencing due to evolutionary time, and pharmaceutical applications.
A hotbed of research lately has been in determining these effects of selection according to evolutionary time. Several approaches for detecting selection have been presented to the scientific world. The codon-degeneracy model (McClellan, 2000) is one of the most powerful approaches. However, given the intense nature of calculations involved in this model, effective applications have been out of reach.
The codon-degeneracy model utilizes information about patterns of codon structure and phylogenetic tree topology to estimate neutral expectations. By comparing the fit of these neutral expectations to inferred nucleotide substitution frequencies it is possible to determine whether selection has occurred.
Creating a computer software program that implements the codon-degeneracy model was a difficult problem due to the complications of adapting the model into something a computer could utilize. This required a background in Statistics, Mathematics, Biology, and Computer Science. The program CDM (Codon-Degeneracy Model) is the result of collaboration with individuals with these backgrounds.
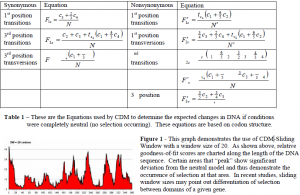
The CDM algorithm is fairly simple. Expected patterns of substitution are estimated by first counting the codon compositions across the sequences of DNA data. Then utilizing the codondegeneracy model-derived equations (Table 1), expected frequencies of nucleotide substitution are calculated. Observed patterns of substitution are then estimated by implementation of baseml, from the PAML suite (Yang, 1996), and comparing DNA sequences to infer substitution events.
The expected and observed patterns of substitution are compared statistically by calculating a goodness-of-fit score between them using a chi-squared distribution. The greater the goodnessof- fit score, the more the data has deviated from neutral expectations, indicating selection. This score is partitioned into synonymous and nonsynonymous substitutions which also allow for measure of substitutional saturation.
The CDM program is a java application which allows for easy accessibility on any computer. It has already been used in research studies (McClellan, 2003) and an article about the program is currently in review in the journal BioInformatics. Interest in CDM allowed for a selected group of BYU students to attend two different conferences; Evolution and Molecular Evolution. During these conferences, much interest was expressed by colleagues from universities all around the country and may lead to future opportunities.
The creation of the CDM program has “opened the door” to many research projects. These projects include studies on genetic codes as evolutionary filters, studies on large data sets to determine areas of selection, and generating data sets on the basis of a neutral model. These areas of research led to another area of need, a sliding window.
During the creation of CDM and due to inspiration from Dr. David McClellan, a sliding window was designed. Since the model of CDM can determine selection on an entire set of molecular data, a sliding window would allow CDM to be broken down to smaller sets within the data and determine localized selection. This would allow the determination of selection all along the gene (Fig. 1).
Future applications of the CDM algorithm are many. They include: reconstruction of phylogenetic tree topologies, testing of more specific neutral null hypotheses, implementation of better statistical tests, and estimating differential selection between active sites and functional domains. Further research in these areas will provide a means to conduct in-depth studies into the relationships, lineage and evolution of organisms of interest.
References
- McClellan, D.A., 2000. The codon-degeneracy model of molecular evolution. J. Mol. Evol., 50, 131-40.
- McClellan, D.A., Whiting, D.G., Christensen, R.C., Sailsbery, J., 2003. Genetic Codes as evolutionary filters: subtle differences in the structure of genetic codes result in significant differences in patterns of nucleotide substation. J. Theoretical Biology.
- Yang, Z., 1999. PAML (Phylogenetc Analysis by Maximum Likelihood), version 2.0, London, UK.