Ashley Nelson and Dr. Keith A. Crandall, Integrative Biology
Introduction
This study of the Oreohelix peripherica land snail is a continuation of a study sponsored last year by ORCA. In 1993, Ecosearch, Inc. suggested that O. p. wasatchensis, located at Rainbow Gardens near Ogden, deserved being upgraded to having an endangered status according to the US Endangered Species Act because of its reduced area of occurrence and the depletion of its habitat. The phylogenetic hypothesis gathered from the data obtained in last year’s study suggests that the Rainbow Gardens samples are not endemic to that area because they are genetically similar to other populations from different localities. Since the population at Rainbow Gardens is not genetically isolated and is related to diverse populations spread over a large geographical area, these data suggest that it is not in need of special conservation efforts. However, this information does not mean that there may be other populations of land snails that may merit an endangered status. Before we can formulate conclusive results, we need to analyze a greater number of specimens from different geographical areas and study the phylogenetic relationships within the Oreohelix species complex.
The land snail Oreohelix species complex consists of 35 species that are distributed throughout the western United States, southwest Canada and northern Mexico. The majority of these species are endemic to the Great Basin and the Rocky Mountains (Bequaert et al., 1973). Pilsbry (1939) made the last taxonomic revision of the Oreohelix snail in Utah. He described seven species: Oreohelix eurekensis, O. haydeni, O. parawanensis, O. peripherica, O. strigosa, O. subrudis, and O. Yavapai. Traditionally, three subspecies have been ascribed to O. peripherica: O. p. wasatchensis, O. p. newcombi and O. p. weberina.
Molecular phylogenetic studies can provide a useful framework for determining the status of the species, in defining conservation efforts, and assess the degree of genetic isolation between conspecific populations (Sites and Crandall, 1997). The goals of this project are to:
1) Collect new data from mitochondrial (12S, 400 bp) and nuclear (ITS, 700 bp) genes to determine the phylogenetic relationships within the land snail Oreohelix peripherica species complex.
2) To assess the population structure and conservation status of O. p. wasatchensis in its range of occurrence.
Materials and Methods
Land Snails were collected to fill the geographic gaps in the phylogenetic analysis of O. peripherica in Utah. We collected samples from Salt Lake, Utah, Wasatch, Summit, Duchesne, Daggett and Uintah Counties. Some Oreohelix strigosa, O. haydeni and O. subrudis samples were also collected in these areas. DNA was extracted using the Salting-Out Method (Crandall et at., 1999). All polymerase chain reactions (PCR) were carried out on a Perkin-Elmer 9600 using these conditions: 5 minutes at 96°C, 50 cycles of 95°C for one minute, 40-55°C for one minute, 72°C for one minute, followed by 72°C for five minutes. These primers were used, 12S F 5’-AGCGACGGGCGATATGTAC-3’ and 12S 1R 5’-GAAACCAGGATTAGATACCC-3’, ITS ETTS1 5’-TGCTTAAGTTCAGCGGGT-3’ and ITS ETTS2 5’-TAACAAGGTTTCCGTAGGTGA-3’. ITS PCRs were cloned using Invitrogen® TOPO cloning kit. Successful ITS clones and 12S PCR products were purified using a GeneClean® II kit (Bio 101). Sequences were obtained on an ABI 377XL automated sequencer using the ABI Big-Dye Ready-Reaction kit using standard cycle sequencing protocol. Phylogenetic relationships were estimated using maximum parsimony (MP), neighbor joining (NJ), and maximum likelihood (ML) methods as implemented in PAUP* 2002 (Felsenstein, 1981). NJ and ML assume a model of evolution; hypotheses relating to the molecular evolution of the nucleotide sequences were tested using Modeltest Version 3.0 (Posada and Crandall, 1998). Confidence in the resulting relationships were assessed using bootstrap analysis (Felsenstein, 1985)
Results and Conclusions
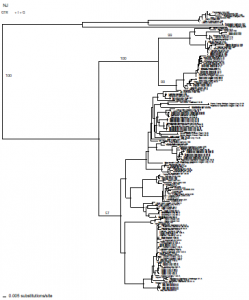
In total, 229 sequences were analyzed using the NJ method for 12s data, assuming the General Time Reversible plus Inversion plus Gamma (GTR + I + G) model of evolution. The phylogenetic relationships inferred from this tree were collaborated with ITS data analyzed using a Bayesian approach. The ITS analysis supported the phylogenetic relationships established with the NJ tree. The species located at Rainbow Gardens is genetically similar to other specimens located across a wide geographical area. Thus, it is unlikely that this species is in need of special endangered status. It has been interesting to learn more about genetic relationships and evolutionary theories. Even though this project has been very challenging, I am satisfied knowing that I have been able to further my education. The numerous lessons I learned, no doubt, will be of a considerable advantage in my future medical career.
References
- Bequaert, J. C., Miller, W. B., 1973. The Mollusks of the Arid Southwest with an Arizona Check List. The Arizona Press, Tucson, Arizona.
- Crandall, K. A., Fetzner, J. W. Jr., Lawler, S. H., Kinnersley, M. and Austin, C. M., 1999. Phylogenetic Relationships among the Australian and New Zealand Genera of Freshwater Crayfishes (Decapoda: Parastacidae). Australian Journal of Zoology 47, 199-214.
- Felsenstein, J., 1981. Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach. Journal of Molecular Evolution 17, 368-376.
- Felsenstein, J., 1985. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 39, 783-791.
- Pilsbry, H. A., 1939. Land Mollusca of North America (North of Mexico). Monograph No 3 of the Academy of Natural Sciences of Philadelphia, Vol. 1, Part 1. Philadelphia, 377 and 573.
- Posada, D., Crandall, K. A., 1998. Modeltest: Testing the Model of DNA Substitution. Bioinformatics 14, 817-818. Sites J. W. Jr., Crandall, K. A., 1997. Testing Species Boundaries in Biodiversity Studies. Conservation Biology 11, 1289-1297.
- Swofford, D. L. 2001. PAUP* Phylogentic Analysis Using Parsimony and other Methods. Sinauer Associates, Sunderland, MA.
