Keith Tanner and Dr. Dennis Shiozawa, Department of Biology
The Great Basin contains many closed basins, separated by north-south lying mountain ranges and low east-west lying divides. The Sierra Nevada, on the western edge of the Great Basin, generates a rain shadow, making Nevada the driest state in the nation, with Utah as the second driest. The dryness of the Great Basin makes water a critical resource for life – for both humans and native organisms. As western urban areas have grown, water has become a limiting resource for both urban and rural development
The Southern Nevada Water Authority (SNWA) plans to pump deep aquifers in isolated basins of eastern Nevada. Springs in these basins are at risk of drying if the water table is lowered. While SNWA will mitigate spring desiccation, it is unknown which invertebrate taxa are capable of recolonizing, nor how unique populations within or between basins are.
We hypothesize that aquatic invertebrates that disperse slowly through water (gastropods, crustaceans) will show genetic distinction between basins and springs within basins. Flying aquatic invertebrates will show intermediate to little genetic distinction between springs within basins due to flight. If this hypothesis is correct, deep aquifer pumping could result in large losses of unique aquatic invertebrates.
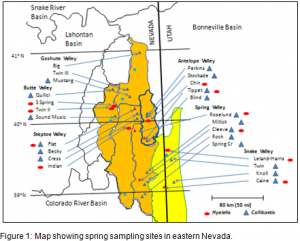
We collected Pyrgulopsis, a native snail and the amphipod Hyalella azteca. Collection occurred in parts of six basins of the Great Basin (see figure 1). We examined eight to ten springs per basin, identifying at least four springs per basin containing Pyrgulopsis and H. azteca.
At each site invertebrates from all taxonomic classes were collected and stored in 95% alcohol. Specimens were counted and identified to species where possible. Later, isolation of DNA was performed on individual organisms using the Puregene DNA Extraction protocol (Evans Lab, BYU). Primers were used to amplify the nuclear 28S ribosomal gene and mitochondrial cytochrome c oxidase subunit I (COI). These genes show evidence of evolution when compared with the same genes of different populations.
Amplified gene sequences of H. azteca were aligned using SequencherTM v4.6 (Gene Codes Corp., Ann Arbor, Michigan). Analysis of the ribosomal 28S gene gave few genotypes, but these data have not yet been completely analyzed. With COI, several hundred individuals were amplified, resulting in 61 haplotypes. Phylogentic trees were constructed using this data. These trees are similar to genealogical trees, with each branch extending to a different ancestor.
These and other data indicate little gene flow among populations of H. azteca. This is according to our hypothesis and tells us that basin drainage connection is limited. Poor gene flow also hints at possibilities of different species within basins. We also found east to west dispersal of H. azteca to be very unique indicating rapid evolution. Therefore if the SNWA chooses to tap these aquifers, endemic species such as H. azteca and many others could vanish.
Due to limits of time and funding, many specimens went unused. Lack of time prevented a larger sample size and processing of any snail samples. The gene amplification protocol proved to be an obstacle also. Other specific nuclear genes had been previously selected but were incompatible with our amplification parameters.
Unfortunately no flying aquatic insects were genetically analyzed. Strong flying insects such as dragonflies, mayflies and caddisflies, could be included in future research. Once analyzed, genes of flying aquatic insect will hopefully show little genetic uniqueness among basin populations due to their strong ability to disperse.
This project was an invaluable addition to my education. It provided an intense view of human and invertebrate interactions within the ecosystem. In addition to the many skills gained, I learned how ethics participates in everyday decision making for both scientists and political leaders.
Special thanks to Dennis Shiozawa, Heather Stutz, Christopher Hansen, Paul Evans, and the ORCA Department at Brigham Young University.
