Robert Kitz and Dr. Sandra Burnett, Microbiology & Molecular Biology
The Macrophage Fas-Induced Apoptosis (Mafia) mouse is a transgenic mouse model designed by Dr. Sandra Burnett to study the role of macrophages in the absence of macrophages. The Mafia transgene contains the genetic code for the Green Florescent Protein, a suicide gene and a c-fms promoter that makes the gene specific to macrophages. The transgene inserted randomly in more than one place causing it to express at different levels within different mice. In order to identify the separate insertion points within the mouse genome, I used several methods of Polymerase Chain Reaction (PCR) testing followed by DNA sequencing.
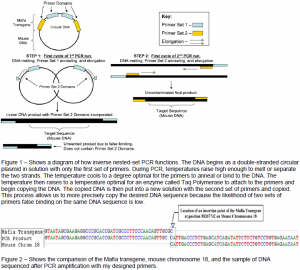
I first started with an inverse nested-set PCR method in order to amplify the unknown flanking sequence to the Mafia transgene using primers developed from the end of the known Mafia sequence (see figure 1). After amplification, I sequenced the PCR product and performed a search on the Genbank database, resulting in a match on the mouse’s 18th chromosome. After developing several more primers to bind on chromosome 18 at continuing lengths downstream from the Mafia transgene, I ran further PCR testing with some of the first primers situated on the Mafia gene. This testing produced DNA amplifications that matched the 18th chromosome at the same location as the first. To verify that the Mafia transgene had inserted at this site, I developed several more primers at increasing lengths on the Mafia gene upstream from the identified sequence on chromosome 18. After PCR testing and DNA sequencing, a sequence containing both Mafia transgene and mouse chromosome 18 was amplified (see figure 2).
The proceeding work resulted in identifying one location of the Mafia transgene inserted backwards at position 58267742 on mus musculus’ chromosome 18. This site is located within the c-fms portion of the chromosome. This is interesting because the Mafia transgene contains the c-fms sequence indicating that there is a possibility that Dr. Burnett unintentionally created a site-directed transgene.
I am continuing to work to validate this insertion site by developing and testing primers that will amplify DNA from a few hundred base pairs upstream from position 58267742 to just a few base pairs onto the end of the backside of the Mafia transgene. Once this amplifies, DNA sequencing should show that the backside of the Mafia transgene is located at position 58267741, just one base pair ahead. This will verify that the transgene did in fact insert backwards between position 58267741 and 58267742. It is also possible that some of the c-fms mouse sequence was removed when the transgene inserted. This will be shown if amplification occurs using primers at a position much further upstream than 58267742.
To further validate the original primers I developed, I will use them to perform PCR tests on additional samples of mouse DNA. If these tests amplify DNA, I will sequence that DNA to confirm these samples also contain the transgene at position 58267742. If the primers are validated in this way, I will be able to use the primers as a genotype test to verify if future mice are Mafia mice.
I put together a poster presenting the questions, methods, and findings of my research. I also prepared an oral presentation explaining the same and present them both at the Autumn Immunology Conference in Chicago in November of 2008.
I was a little surprised and excited to find that there were many researchers who found interest in my project. A few approached me who had attempted to locate the insertion points of a transgene they are working with. One researcher was not successful in identifying her transgene location. We were able to discuss what might have gone wrong and further steps she could take to be more successful. I hope to publish these findings in an academic journal so that others, too, may use it to assist their research.