Benjamin L. King and Dr. Peter J. Maughan, Plant and Animal Sciences
DNA barcoding is one of the most important, novel techniques to be introduced in molecular systematics and genetics research in the last decade. Barcoding has been established as a consistent and reliable indicator of taxonomic relationships in eukaryotes, particularly in animals, where a uniform barcode sequence in the mitochondrial genome is used. In plants, a standard barcoding system has only been settled upon in the past year, and it is based on combining DNA sequences from three regions of the chloroplast genome. As part of the greater C. quinoa project overseen by the faculty of the Plant and Wildlife Sciences Department, my project focused primarily on the characterization of one of the three barcode sequences – the trnH-psbA spacer – within the group of Chenopodium (goosefoot) species of North and South America.
The barcoding project has at its roots two main objectives. First, we needed to verify the validity of barcoding assays in this little-studied genus. The region we chose, which had been proposed in the literature, was the trnH-psbA intergenic spacer (IGS). Being a part of the chloroplast genome, this sequence is uniparentally inherited only from the female plant (maternally). This uniparental inheritance is an important qualification for barcoding sequences, as it reduces or eliminates the interference of such phenomena as recombination, which might otherwise confound the phylogenetic analysis that is central to DNA barcoding purposes.
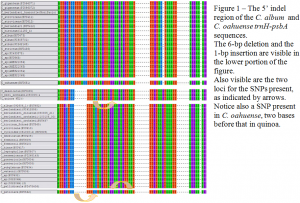
The trnH-psbA spacer analysis included 6 S. American, 16 N. American, 1 Hawaiian, and 6 Eurasian Chenopodium taxa. Each species was represented by multiple tissue and DNA samples from a total of 89 individual accessions, or an average of ~3 samples per species. The data accumulated from the chenopod collection revealed several important identifying characteristics, including a region which contains two insertion-deletion (indel) events: a 6-bp deletion and a 1-bp insertion – that have proven useful in our analyses. These sequencing data suggest a close phylogenetic relationship among some N. and S. American diploids and allotetraploids, including quinoa (C. quinoa), and suggest a polyphyletic origin of the allotetraploid C. berlandieri complex, possibly including C. quinoa.
There were several surprises in this project. Based on the assumptions of traditional taxonomy we expected to find a close molecular relationship between C. oahuense, the only chenopod species endemic to Hawaii, and several North American species. However, the indel region referenced above indicates a closer relationship for C. oahuense with the Eurasian C. album complex than with the North American group. This suggests a possible trans-Pacific migration from Asia sometime in the evolutionary history of the species.
There were also several important polymorphisms within the sequence, although on the whole sequence identity was very high. For instance, Figure 1 shows a portion of the indel region mentioned above, where the North and South American group show a >99% homology. However, this group can be further subdivided into three classes: quinoa and identical sequences, which have a single nucleotide polymorphism (SNP) relative to the Old World stock (along with the indel previously mentioned); diploid and tetraploid accessions with the Eurasian version of the SNP (along with the indel); and a Chilean accession of C. petiolare, which has two SNPs relative to quinoa (arrows indicate locations of SNPs).
One of the greatest difficulties we faced in performing the assays appropriately involved the taxonomic identification of the plants in question. Chenopodium is a highly morphologically diverse genus, and is poorly characterized by taxonomists. There also seems to be a high level of heteromorphy combined with phenotypic plasticity in most of these species, which further confounds classification within the genus. Several species have recently been reassigned to other genera based on phylogenetic analysis, and were excluded from the final analysis of our data. Our conclusions support this reclassification, and also suggest the possibility of further reclassification. For example, C. album and C. berlandieri, in particular, have served as “catch-all” taxa for grouping accessions that do not fit into the other species designations. Both are notoriously polymorphic and are highly invasive weeds that have spread trans-oceanically. After sequencing, it became clear that many of the accessions tentatively classified as these two species should be reclassified. The indel we discovered should prove valuable for this purpose.
This project is the first step toward more rigorous standards of classification based on both morphological and molecular structure. Further research on other important sequences will clarify relationships between the species of the genus and allow a more accurate phylogenetic tree to be developed.