Rebecca Carlyon and Dr. Joel Griffitts, Department of Molecular and Microbiology
Symbiosis is the process by which two different species interact in nature. Relationships can be either mutualistic, where both species benefit from the situation, or parasitic, where only one organism benefits. Shinorhizobium meliloti is a nitrogen-fixing soil bacterium which is involved in a mutualistic symbiotic relationship with alfalfa and other compatible host plants. The plant allows the bacterium to colonize the plant in forming a root nodule. This symbiosis provides the bacteria with carbon resources, and the plant with enough fixed nitrogen to flourish in the soil.
How does this infection process occur? There exists a process in which a bacterium receives a signal from the environment which starts a series of events which initiates infection. This process is vital for understanding both disease-causing and beneficial bacteria, and could prove to be vital for the agricultural process.
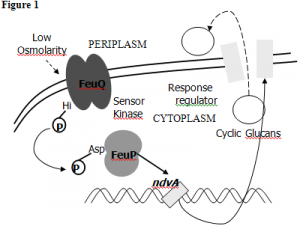
In the genome of S. meliloti, there is a three gene operon consisting of feuN, feuP, and feuQ. These genes are all co-transcribed, and conserved in other host-associated alpha-proteobacteria. In our lab, we have previously discovered the roles of FeuP and FeuQ, which comprise a two-component system (see Figure 1). FeuQ is a sensor kinase which is responsible for sensing the environment for signals, and then forwarding it to FeuP, a response regulator which acts as a transcription factor. They positively regulate the expression of ndvA, which is known to make a cyclic glucan exporter. Cyclic glucans are polysaccharides that are important for various host-bacterium interactions. Without the FeuP/Q two-component system, infection can not occur.
The novel protein, FeuN, is located on the same operon as FeuP and FeuQ. FeuN negatively modulates signaling through the FeuP/Q two-component system. With over-expression of feuN, ndvA expression decreases. Interestingly, deletion of feuN is lethal. This lethality is suppressed in a feuQ deletion mutant, underscoring the functional interaction between these genes.
The purpose of my project was to describe the molecular interaction between FeuP/Q/N and to understand what FeuN does to the two-component system to negatively regulate it. For convenience, the FeuP/FeuQ pathway was moved to E. coli. Through a number of tests, it was found that the E. coli pathway works similar to the native pathway. Using the E. coli system, I designed a strategy where I was able to find variants of FeuP/Q that don’t respond to the inhibitory effects of FeuN.
In this screen I first mutagenized the feuP/Q genes and digested them into a plasmid with antibiotic selection. This plasmid was then transformed into an E. coli strain that has two other plasmids. The first was feuN, and the second was a reporter plasmid with ndvA promoter fused to a lacZ gene. This second plasmid allowed us to test the level of feuN inhibition on the pathway. Once all three plasmids were in the E. coli strain, I measured the levels of ndvA expression. I was looking for a mutation that reads the same ndvA expression in E. coli as when feuN is not present.
The screen first consisted of plating the three plasmid system in E. coli onto LB-Tc, Km, Ap, Arabinose, X-gal plates. This allowed me to look for the blue phenotype that is expected of a variant that doesn’t respond to the inhibitory effects of FeuN. Unfortunately, this phenotype was also consistent with a loss of function FeuQ phenotype. Subsequently, I had to perform a quantitative analysis by doing a B-galactosidase assay. Once I had confirmed the strains for the correct phenotype, I then sequenced the strain using four different sequencing reactions since the feuP/Q genes are quite long.
Of the thousands of colonies that I screened through, only 20 were strains that didn’t respond to FeuN inhibition. These strains were very informative in that even though we allowed for both FeuP and FeuQ mutants, only FeuQ mutants turned out to have the loss of FeuN inhibition phenotype. This suggests that FeuN interacts with FeuQ to inhibit the pathway. We also learned FeuN must interact with FeuQ on its transmembrane domain which also suggests the localization of FeuN. Further analysis will allow us to understand how FeuN negatively regulates the two-component system, but this analysis provides a valuable start.
