Nick Candelaria
Each year, more than 170,000 women are diagnosed with breast cancer in the United States alone [1]. The risk for developing breast cancer can largely be attributed to the levels of exogenous and endogenous hormones in the body [2]. A hormone known to have drastic effect on cancer cell proliferation is estrogen, a regulator of cardiovascular, bone, and reproductive systems [3]. Estrogen is produced endogenously in the granulosa and theca cells of the ovary, and is found in a variety of forms: 17-estradiol, estrone, and estriol. These estrogen subtypes are derived from cholesterol, and contain varying isoforms of 18 carbons [4]. Furthermore, estrogens can also be obtained from a variety of exogenous sources. Phytoestrogens are obtained in diet, are compounds with estrogenic activity and are found in soy, fruit, vegetables, seeds, and nuts. Some well-characterized phytoestrogens are genestien, glycitein, daidzein, enterodiol, and enterolactone [6].
Once in the cell, estrogen receptor modulates gene expression by binding estrogen, which subsequently binds to DNA as a cis-regulatory element and alters gene expression [4,5]. A type of estrogen receptor, ER, belongs to the nuclear receptor superfamily. ER is 66 kDa, is isolated to the nucleus, and is widely distributed in the epidydimus, pituitary, testis, ovary, kidney, uterus, and adrenal [4,6]. Because another type of estrogen receptor is not expressed at high levels in breast tissue, only ER will be studied.
Determining Estrogen Receptor a (ERa) Recruitment to Downregulated Target Genes when Bound with plant phytoestrogen ligands
After estrogen receptor binds to DNA, its specific effect on gene expression depends on the type of estrogen bound to estrogen receptor, the characteristics of the region surrounding the promoter, and the addition of coregulators (co-repressors/co-activators) which function to precisely alter transcriptional response [6]. Until recently, it was not known that ERa could participate directly in transcriptional repression of target genes. However, a genome-wide study mapping ERa binding sites identified 286 high-confidence ER binding sites, and of those, 116 are targets for genes that are directly down-regulated by ER by an unknown mechanism [8]. Furthermore, our lab has worked with the following co-repressors bound to estrogen receptor at the binding sites of several down-regulated genes: NCoR, NRIP1, and SMRT. A graduate student in our lab performed chromatin immunoprecipitation experiments to determine the binding order of ER, co-repressors, co-activators, and promoter transcriptional machinery. He was also successful in detecting necessary histone modifications made by co-repressors bound to ER at the promoters of these down-regulated target genes. However, his study only looked at treatment using estradiol, and more work needs to be done to characterize transcriptional repression by ER when dietary estrogens (genistein/resveratrol) are bound.
Chromatin immunoprecipitation (ChIP) is a method used to quantify recruitment levels of a DNA-bound protein. MCF-7 cells (breast cancer cells) were stimulated with the following estrogens: estradiol, genistein, resveratrol, and vehicle (EtOH). Then, segments of DNA segments of interest are immunoprecipitated then quantified using real-time PCR. Mastering protocols for ChIP, tissue culture, and real-time PCR have proven very difficult. Eventually, however, the following results indicate that ER binding adjacent to downregulated target genes could be detected.
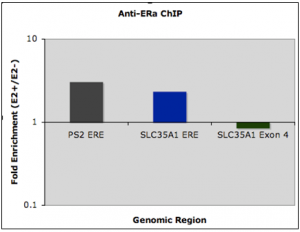
PS2 is a well-studied upregulated target of ER (purpose is a control), while SLC35A1 is a downregulated target gene of ER. Estrogen response element (or ERE) refers to the genomic region where ER binds, and isn’t the expression of the gene itself. (For this reason, fold changes are referred to as “recruitment” to avoid confusion). The SLC35A1 exon 4 is a negative control, and should show no difference in binding to ER. Because the protocol has been established, quantification of corepressor/coactivator recruitment based on phytoestrogen stimulation is possible at downregulated target genes.
As mentioned in the introduction, we were interested in quantifying the amount of protein binding to downregulated target genes using ligands of ER that are not estradiol (such as genistein or resveratrol). First, we needed to look at downregulation of target genes in cell lines expression estrogen receptor to see if that behavior differs when a different estrogen is used. The two cell lines used are T47D and ZR75-1, also estrogen receptor positive breast cancer cell lines. Here is what we saw in those:
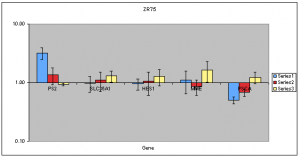
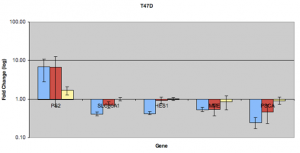
In this case, the blue refers to estradiol treatment. Red refers to treatment with genistein and yellow refers to resveratrol treatment. These can now be overlayed on future ChIP experiments used to quantify binding at these downregulated target genes. The data show that ZR751 cells tend to upregulate normally downregulated target genes when resveratrol is used. In T47D, PSCA, MME, and SLC35A1 appear to be able to downregulate when genestien is used. HES1, however, does not appear to respond in a similar manner. Note: a replicate issue on genstein/PS2 will have to be addressed at some point in the future. An interesting and unexpected conclusion that could possibly be drawn from the following data is this: (a) resveratrol does not appear to be as strong an estrogen as some have supposed, or (b) resveratrol appears to be able to modestly upregulate target genes of estrogen receptor, but prevents downregulation by some unknown mechanism. Therefore, the ChIP study is needed to confirm this. If this turns out to be the case, it may provide some very interesting information to those interested in foods that may help in the prevention and fight of breast cancer.
RNP-IP Portion
Chromatin immunoprecipitation is a technique used to quantify binding of a protein to DNA. Proteins are crosslinked using formaldehyde to DNA. Once the proteins are immunoprecipitated and everything unrelated is washed away, the crosslinking is reversed, and the DNA is quantified used real-time PCR.
Long-non-protein-coding RNAs are RNAs that have diverse activities. One such lncRNA is SRA or steroid RNA co-activator. This lncRNA can bind SRC-1, a protein involved in estrogen response and can help upregulate a gene [9]. We were interested to see if the same crosslinking principles could be employed to precipitate bound lncRNAs from proteins of interest. For that reason, we utilized a relatively new and untested technique called ribonucleoprotein immunoprecipitation to quantify bound RNAs. Early attempts began in May, and were failures, largely because they were used our existing ChIP protocol modified at the last step to purify RNA instead of DNA. One early result led us to believe that our RNA could be isolated at incredible purity. However, attempts at replicating this failed. For that reason, we looked at melting curves and we mapped the primers of our collaborators. Upon doing this, we found that the primers used to study SRA in another paper had a product length well outside of the specs of the PCR machine. We redesigned the primers, and retried the reaction, and saw no difference. We then tried to utilize a RIP protocol that had already been optimized by others to study SRA [10]. Early attempts tried to use our existing protocol. After other failures, we tried their technique in full: here are the results. (we were resisting this technique because it did not include a nuclear isolation step. We thought that we would try the cytoplasmic lysis first, then see if we could isolate the nucleus later).
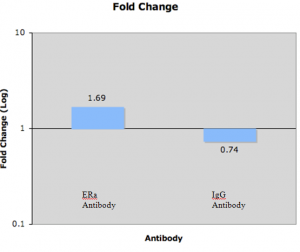
IgG is nonspecific and should show no fold change. Anti-ERa should show an enrichment in SRA binding:
LNC RNA Portion
(This was the original subject of my ORCA proposal: refer to original proposal for background information and plan).
We eventually put a stop to general lncRNA study because our collaborators felt like primer design issues could not be addressed at this point in time. For that reason, only gene expression analyses were performed. A compilation of three biological replicates of each lncRNA was established.
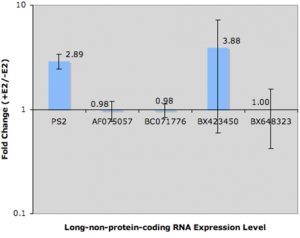
PS2 is the upregulation control. Three of the results for lncRNAs AF075057, BC071776, and BX648323 clearly show no fold change by Estrogen treatment. That was one of the early requirements when deciding which lncRNAs we would be further studied. BX423450 showed similar results for two of the three biological replicates. For some reason, the third biological replicate showed significant upregulation. This could be because of a number of factors: among them, (a) poor RNA extraction, (b) cell handling inconsistencies, (c) or problems with reverse transcription. The possibilities are endless, but because we did three biological replicates, and because the error bar spans both sides of 1, we were forced to conclude that the replicate was an isolated incident (which we were unable to attribute a cause). For that reason, none of the lncRNAs I chose to study in this portion appeared to be regulated by estrogen treatment.
Conclusion
Now that we are making progress on all of these fronts as a lab, we are better prepared to perform the “meta-analysis” of estrogen receptor function in breast cancer cells. We are better able to analyze gene expression, quantify binding of protein to DNA, and quantify binding of RNA to proteins of interest. Now we need to implement these techniques on a wider scale to assemble a better picture of how estrogen receptor can aggravate malignant breast tissue. I would like to thank the ORCA office for their kind donations. This allowed me to stay at BYU over the summer to conduct this research. Again I am grateful for your kind donation.
Sources
- Jemal A, Siegel R, et al. Cancer statistics, 2007. CA Cancer J Clin 2007;57;43-66.
- Monninkhof EM, Elias SG, et al. Physical activity and breast cancer: A systematic review. Epidemiology 2007;18;137-57.
- Lin CY, Vega VB, et al. Whole genome cartography of estrogen receptor binding sites. Plos Genetics 2007; 3, 867-85.
- Gruber CJ, Tschugguel W, et al. Production and actions of estrogens. N Engl J Med 2002;346;340-52.
- Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol 2004; 66;315-60.
- Pearce ST, Jordan VC. The biological role of estrogen receptors and in cancer. Crit Rev Onc Hem 2004;50;3-22.
- Frasor J, Fabio S. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res 2004;64;1522-33.
- Lin CY, Strom A, et al. Discovery of estrogen receptor target genes and response elements in breast tumor cells. Genome Biol 2004; 5;R66.
- Lanz RB, McKenna NJ, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999; 97;17-27.
- Xu B, Yang WH, et al. Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol Cell Biol 2009; 29; 1719-1734.
