Jennifer L. Potter and Dr. Laura C. Bridgewater, Zoology
The human skeletal structure is formed first by cartilage which later ossifies to bone. It follows, therefore, that if cartilage formation is aberrant, the resulting bone formation will be affected. Cartilage is composed of fibrils consisting of three types of collagen: type II, type IX, and type XI. These fibrils are only produced by chondrocyte (cartilage) cells. I was interested in the Col11a2 gene that codes for a subunit in type XI collagen. This gene was found to play a crucial role in cartilage formation. Mutations in the Col11a2 gene can cause skeletal deformities, cleft palate, hearing loss, osteoarthiritis, dwarfism, and even perinatal lethal chondrodysplasia. Due to the fact that mutations in this gene can cause such a wide range and severity of defects, it is important to figure out how this gene is regulated in chondrocytes. Enhancers, often located in the 5’ region of a gene, regulate when genes are turned on and off in a cell and how actively they are expressed. There are two types of enhancers, general and specific. General enhancers control genes in a number of different cell types. However, specific enhancers control certain genes only in a specific cell type. We know that the Col11a2 gene must be regulated by specific enhancers because it is only expressed in chondrocyte cells. Preliminary work has identified two cellspecific enhancers in the 5’ region; however, it also revealed that these were not the only enhancers that controlled Col11a2 expression.1 As such, I was interested in looking systematically through the 5’ region of Col11a2 to find the remaining cell-specific enhancers. By doing this we would gain a better understanding of the regulation of Col11a2, an important gene that when mutated can cause severe deformities. We would also obtain a better understanding of how tissue-specific regulation works in cartilage. This information could then be applied to other genes in different tissues.
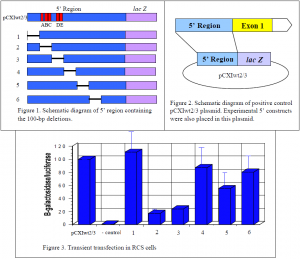
The goal of this project was to thoroughly and systematically search through the 5’ region of the Col11a2 gene to identify cell-specific enhancers. The design of this experiment was to recreate the 5’ region of the gene while deleting 100 base pair (bp) regions of the gene. Jim Clarke, a lab associate, and I created six 100-bp deletion constructs (Fig. 1) with the idea that if there is an enhancer in the missing 100-bp region, the activity of the gene will decrease because the enhancer is no longer present. To measure the reduction in activity from the experimental constructs, a positive control, which contains the complete 5’ region, was used as a standard in the experiments. If the experimental constructs reveal no decrease in activity it can be assumed there is nothing of importance in the missing 100-bp region. However, if a decrease in activity is detected it indicates the presence of an enhancer element in the region that was deleted. Further investigation would then be done to narrow down the precise location of the enhancer element.
The experimental 5’ region constructs were attached to the reporter gene lac Z, which produces b-galactosidase, in the pCXIwt2/3 plasmid (Fig. 2). The experimental plasmids were then transiently transfected into tissue cultures of rat chondrosarcoma (RCS) cells. The amount of b- galactosidase produced was then measured because its expression level reflects the activity of the attached enhancer elements. For an internal control to measure the efficiency of transfection, the pGL2-control plasmid was cotransfected in every reaction. This plasmid constitutively expresses luciferase. Measurements were reported in b-galactosidase units per luciferase units. Each experiment was done five times and normalized to 100% activity in the pCXIwt2/3 control. This allowed for comparison and standardization of the parallel experiments.
The two cell-specific enhancers that are already known are labeled A/B/C and D/E and are located in the 2nd and 3rd deletion constructs (Fig. 1). As seen in figure 3, deletion of these enhancers resulted in a great reduction of gene activity verifying the validity of this experimental approach. However, deletion of the first 100-bp region does not decrease activity at all, indicating there is nothing of importance in that region. The 4th and 6th deletion constructs show a slight decrease in activity, but not nearly as much as the 5th deletion. The 5th deletion showed an approximately 50% decrease in gene activity. This indicates the likely presence of an enhancer element in that particular 100-bp region. Work is currently underway to narrow down the region and to obtain the precise location of the enhancer element. This work has shown that indeed the two known cell-specific enhancers were not the only ones controlling Col11a2 gene expression.
1. Bridgewater, L.C., Lefebvre, V., and de Crombrugghe, B. (1998) Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J. Biol. Chem. 273:143998-15006.