Hyrum Eddington and John M. Chaston, Plant & Wildlife Sciences
Introduction:
Autism Spectrum Disorder is a neurodevelopmental condition which significantly affects an individual’s social behavior and ability to function in daily living. Although there is a genetic component to the disorder, current research suggests that there are other factors that influence the onset of the disease. One of these factors is the gut microbiota of the individual. Because the human microbiome is a complex system that significantly varies from person to person, finding microbial markers indicative of certain disease can be challenging and problematic. Currently, there are no established protocols that can diagnosis an individual with autism on any criteria save their behavior, meaning that the youngest age at which a child can be diagnosed is about 2 years of age. In addition, so far the majority of this research has shown only correlative links between autistic individuals and the microbiome, with the mechanism of how the microbiota might affect the brain to induce such symptoms requiring much elucidation. In performing this study, we hoped to add to or corroborate the findings of other studies that are similarly attempting to demonstrate the presence of significant microbial markers in ASD individuals.
Methodology:
Fecal samples were collected from individuals who have an older sibling with autism but who themselves have not yet been diagnosed with autism. All of these individuals were about or under two years of age. Samples were sequenced using Illumina sequencing technology and PCR which amplifies a region of DNA in the sample which can used to distinguish between different bacteria. Once the samples were sequenced QIIME bioinformatics software was used to determine the taxonomic identity of each isolated DNA sequence. Principle coordinate analysis (PCoA) was used to evaluate the net difference in microbiota composition between our experimental and control populations. ANCOM analysis was then used to identify specific taxa that had significantly different levels of abundance in our at-risk population.
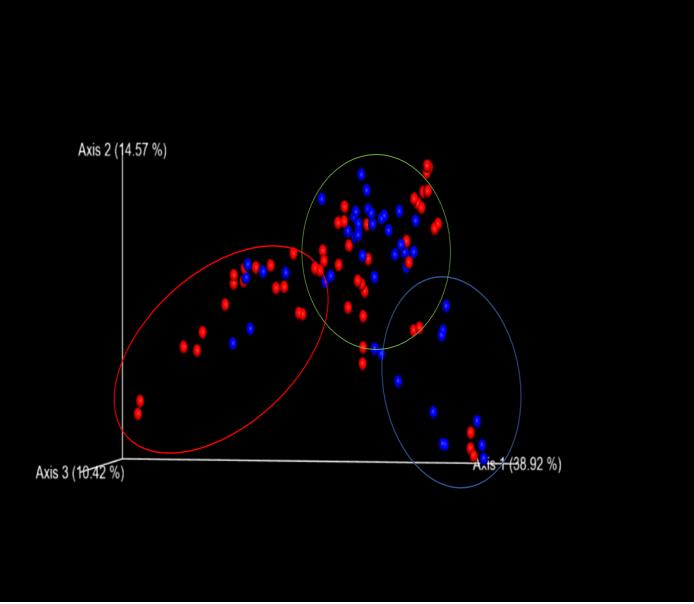
Fig. 1: PCoA shows segregation of some experimental samples (blue) from controls (red).
Results:
Using QIIME bioinformatics software and R statistical analysis, we found that the microbiota of the at-risk children differed significantly from the control population (p- value .008). We also found that the taxa B. fragilis, a bacterium which other literature has shown to be successful at treating ASD symptoms, was significantly lower in abundance in our at-risk population (p-value 2.75e-7).
Fig. 2: Relative abundance of taxa at the family level. Most abundant groups include Lachnospiraceae (brown). Bacteroidaceae is differentially abundant.
Discussion:
B. fragilis has previously been demonstrated to be an effective probiotic treatment for ASD (Hsiao). Treatment with B. fragilis is able to restore the relative abundance of the naturally occurring microbiota and reduce ASD and anxiety-like behaviors in mouse models. However, thus far its promise as a diagnostic marker has not yet been explored. Because current clinical procedures rely heavily on behavioral symptoms to diagnose ASD, a cohort of genetic, microbial, and/or metabolic markers would be extremely useful in standardizing the ASD diagnostic process, as well as elucidating the pathological mechanism by which ASD manifests itself in individuals. Research has shown some success in isolating genetic variables related to autism, however, the complexity of the microbiome makes any predictive correlation of microbial markers with human disease difficult to justify. Our findings corroborate Hsaio’s conclusion that B. fragilis is in some way linked with ASD pathology, and additionally suggests that it may also be possible diagnostic marker for ASD.
Conclusion:
Although many previous studies have studied the effects of the microbiota on ASD individuals, our study was unique in that it showed the possibility of certain bacterial trends in the microbiota of at-risk individuals. Due to funding limitations, we are unable at this time to follow up with the families of these children to asses which of them have received an ASD diagnosis. Such studies in the future would be able to evaluate the predictive power of the possible biomarkers found in this study. Despite this, our findings do suggest that even at-risk individuals who have a genetic predisposition to ASD display identifying trends in their gut microbiota, specifically an extremely low abundance of B. fragilis. Although further research is needed to corroborate this finding, it may prove to be a useful for ASD screening in early childhood.
1. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013; 155: 1451-1463 doi:10.1016/ j.cell.2013.11.024