Taylor Brown, Dr. Chantel Sloan, Department of Health Science
Respiratory Syncytial Virus (RSV) infects almost all infants before their second birthday and can progress to lower respiratory infections such as bronchiolitis or pneumonia. However, few are aware of the effect this has on adults, particularly those who are immunocompromised through organ or bone marrow transplants, treatments for multiple sclerosis (MS), etc. Infection is the leading cause of morbidity and mortality among the immunocompromised. This is of note because RSV is one of the few diseases for which humans are the reservoir species, meaning that this virus only infects and lives in humans. The study investigated genetic susceptibility for a rate of coinfection of respiratory syncytial virus and influenza. The gene investigated was the Mx1 gene. The Mx1 gene has been linked to increased immunity against H5N1 influenza strain in mice and shows promised resilience to respiratory illness when present in humans.
The genetic information for the study was accessed using BioVU, a DNA biorepository linked to de-identified medical records housed at Vanderbilt University Medical Center (VUMC). The study population consisted of adults of all ethnicities, aged 18 and older, with a positive diagnosis for multiple sclerosis (MS). This population was chosen due to the treatment that accompanies MS which essentially eliminates the individual’s immune system. We used an algorithm of ICD 9 and 10 codes known to well identify persons with RSV and influenza. The primary codes used are 487.0 – 487.8 (influenza) and 079.6 for RSV. We used these individuals in the BioVu system and confirmed coinfection by selecting for individuals with a positive test for both RSV and another respiratory infection. Controls were designated by a positive diagnosis for RSV or influenza without a secondary infection identified on the panel.
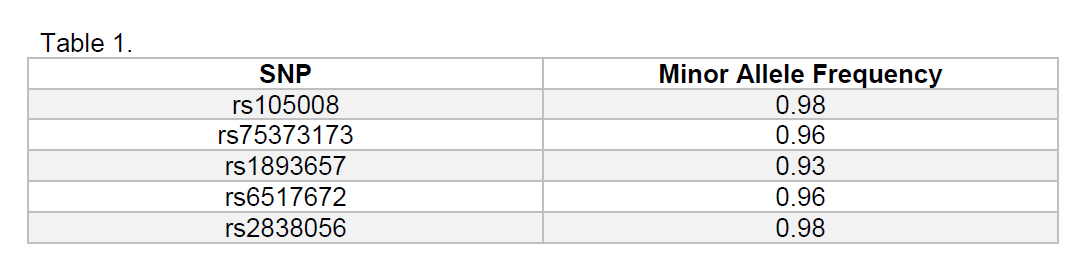
The primary analytical method used was linear disequilibrium and calculated minor allele frequency to measure variation. The MX1 gene was broken down into single nucleotide polymorphisms or SNPs. SNPs are locations of genetic material on the gene that code for a specific trait. In this case we are looking for any SNPs that appear significantly more often, meaning it is not coincidence that those infected have the same genetic markers. SNPs are often different from the normal or “major” allele by only one or two base pairs out of hundreds or thousands. The following table shows the top five SNPs showing significant abnormality resulting from our analysis:
All the data were collected and displayed in figure 1. These SNPs from the table show the most likely tendency to be found in the populations of MS with respiratory co-infection with RSV. Upon review of the literature, there is no specific mention of these SNPs referenced, however further research is being conducted on MX1. Certain SNPs have been looked at in other diseases such as cancers or HIV, but no in-depth research has been done concerning resistance to RSV.
In order to fully use the potential gene markers we have discovered, more research must be done so we may more fully understand not just MS populations, but all other immunocompromised adults. Further study of the MX1 gene could have an impact on HIV positive patients, cancer treatments, and many other diseases and infections that impact the immune system.
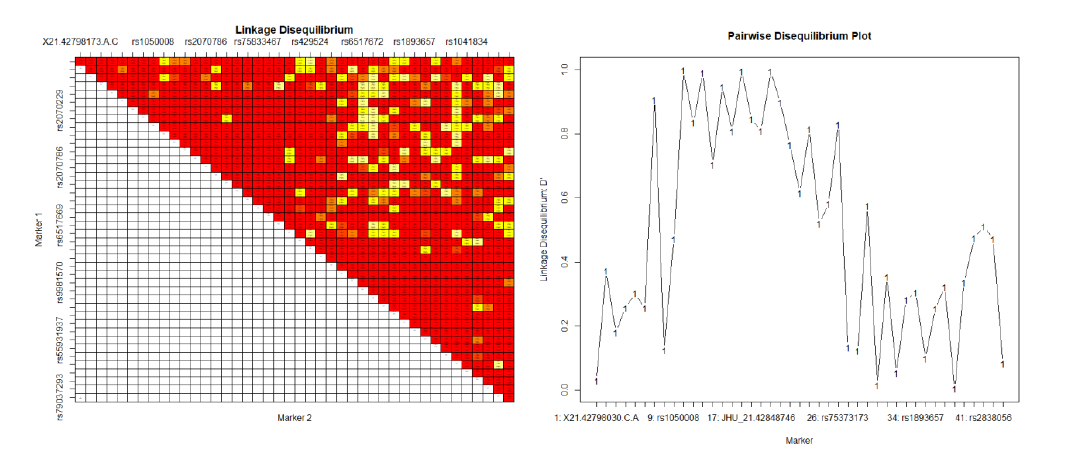
Figure 1 – The first graph is the Linkage Disequilibrium Matrix, showing which SNPs are often found on the same allele within the MX1 gene. The dark red shows a randomization, and the lighter color shows a significant probability that the genes are linked. The graph on the right is the Pairwise Disequilibrium Plot, showing a certain SNP and showing how significant the pairing of that SNP is to other SNPs.