Mark Roth and Dr. Jonathan Alder, Department of Physiology and Developmental Biology
Conditional cell lines are widely used throughout biochemical research. The basic concept is that you can delete a gene you are studying upon the addition of a drug, to cells in culture, and then use the knockout cells to study the genes effects. This process has been facilitated by the advent of the CRISPR/Cas9 system that allows a broad range of genome editing in living cells. Cas9 introduces a double strand break in DNA at a site specified by the CRISPR (guide RNA), and by changing the CRISPR sequence researchers can introduce these double strand breaks almost wherever they want. This in turn allows them to take advantage of a specific repair mechanism, called Homologous Recombination) to make changes to the DNA. The problem is that this repair pathway is rare and often outcompeted by the less reliable mutation prone Nonhomologous end joining (NHEJ). By creating a new construct, we can insert modified genes into the genome in an already reliable method, called Flipping In, that will allow us to then take advantage of cells’ more common repair method, NHEJ, to delete the endogenous version of our gene of interest. Finally, because of the construct we created we can delete our modified gene upon the addition of a drug. In this method we create a reliable method to create conditional genes that does not rely on a rare repair pathway.
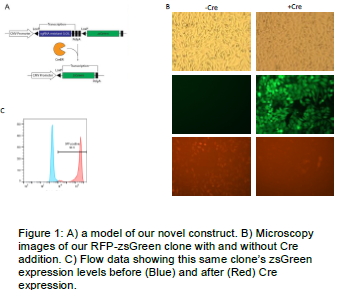
Our goal was to create a novel construct to introduce our desired conditional gene. This method would simplify the genome editing that was necessary, by allowing us to do more molecular biology and less in vivo genome editing. The plan was to create a plasmid construct that could be inserted into the genome using the Flip Recombinase system. Our plasmid construct would have an easily edited site flanked by two LoxP sites (recognized by Cre recombinase that can be used to delete DNA between LoxP sites). Downstream of this site and before the 3’ LoxP site we inserted three ribosome dissociation regions to prevent further transcription. After the 3’ LoxP site we inserted the zsGreen gene. The theory is that when your gene is being expressed zsGreen isn’t When you add Cre Recombinase your gene is excluded from the genome and zsGreen is transcribed instead, acting as a reporter of successful deletion (FIGURE 1A).
To test the success of our construct we introduced RFP into the MCS using a Gibson assembly. Using Flip recombinase, we introduced the purified plasmid into the genome of HEK293 cells. After selection, we observed almost 100% RFP expression and very little zsGreen expression (FIGURE 1B). Upon the addition of Cre Recombinase these conditions switched and we saw little residual RFP expression and almost 100% GFP expression (FIGURE 1B-C).
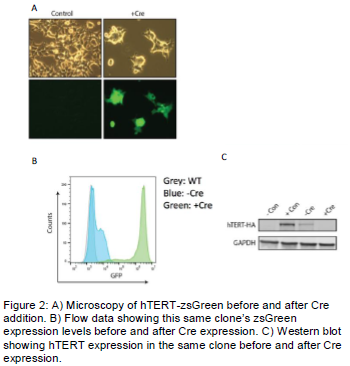
To Further validate this construct, we tested it with hTERT. This construct works in conjunction with the CRISPR/Cas9 system. By engineering a CRISPR resistant version of hTERT, we were able to delete the endogenous copy of hTERT using a CRISPR targeting the catalytic region of hTERT, after the introduction of our construct into the genome. This effectively made or cell line dependent on our exogenous copy of hTERT. This was confirmed by sequencing. We used microscopy, Flow cytometry, and Western blotting to confirm the presence of hTERT prior to Cre addition and its absence after Cre addition (FIGURE 2).
We were able to create a novel tool for use in creating conditional cell lines that we validated with a number of different methods including microscopy, flow cytometry, and western blotting. This construct allows the conditional deletion of your gene (upon the addition of Cre), which allows you to study genes vital to a cell’s survivability, or grow up large numbers of cells and simultaneously delete the gene in all of them.
