PJ Tatlow and Faculty Mentor Stephen Piccolo, Department of Biology
Over the course of the past year I have been able to put a lot of work into creating a tool for scientists, those with computational background and without, that provides a simple web interface for downloading data from large, publically available datasets. It allows users to select a dataset that we have pre-processed, enter filters based on the samples they want to research, and download only that portion of the data. This will save researchers a lot of time, as well as reduce their reliance on bioinformaticians. Because of the increasing number of biological datasets that are becoming publically available, we hope this tool will facilitate more scientists taking advantage of the wealth of data that is now at their fingertips.
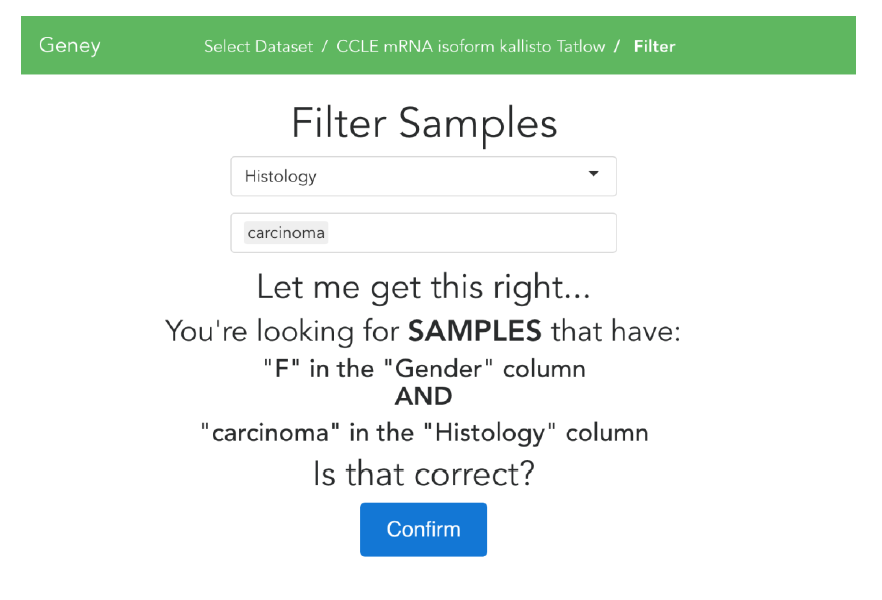
We’ve decided to name the tool Geney, and the code for the site can be found on GitHub (https://github.com/srp33/Geney). I’ve written the front-end web application using a modern JavaScript framework called Vue, and the back-end server in Python. Currently, the site’s workflow is very simple. On the first page, there are a list of datasets, and users can search and sort them to find the ones they’re looking for. Once they’ve found the dataset their interested in, they can select it and input as many or as few filters as they’d like. These filters allow them to pick the samples that match whichever criteria is best for their research (i.e. female patients with carcinoma). Below is a screenshot of the filters page.
Once they’ve selected filters that give them the samples they’re interested in, they go to the downloads page where they can select the genes that they want to study, the sample variables to include in their download, and finally download the data. The Python server then downloads just the samples and variables they requested in the file format of their choosing (TSV, CSV, or JSON). We have also integrated Geney with a third-party visualization tool called Plot.ly (https://plot.ly), a very powerful plotting tool that can create pretty much any type of chart and export images. This pairing should allow users to not only analyze data easier, but also show their discoveries through high quality and easy to create plots.
But not only are people going to be able to use Geney on our servers with the datasets we’ve prepared, but I’ve developed it in a way that will make it easy for others to bring up their own Geney server. Many applications that focus on working with big data require large-scale distributed databases that require lots of management and large servers. We decided to take a federated approach, so that the load can be shared across a variety of web-servers hosted by different groups rather than one very expensive site. Additionally, if scientists have private data that they cannot share they can host their own private version of Geney to allow easy internal access. Because of this federated architecture, we needed the datasets to be highly portable. To do this, we researched many different embedded data formats that allow fast searching access, as well as created a pipeline which allows datasets to be created and put into the correct Geney format. We call this pipeline WishBuilder (https://srp33.github.io/WishBuilder), and we’ve already prepared over 50 datasets for people to use with Geney, with many more to come!