Nate Batey, Zach Frederich, Morgan Fronk, Nate Jenkins and Jonathon Hill, Physiology and Developmental Biology
Introduction
Congenital Heart Disease (CHD) is the most common type of birth defect in the United States,
causing more first-year infant deaths than any other birth defect (1). While the causes of CHD
are varied, genetic defects are known to play a significant role. Identifying and studying genes
involved in embryonic heart development can enhance understanding of the genetic factors in
CHDs. The camta1 gene is a promising candidate due to its transient expression in heart
morphogenesis and its relationship to camta2 , the only other member of its family in vertebrates
and a known player in adult heart failure (2,3).
Methodology
In order to determine the function of camta1 , we have been using CRISPR-associated protein 9
nuclease (CRISPR/Cas9) to microinject zebrafish embryos to create mutants for observation.
Cas9 is a protein that cuts DNA at a sequence designated by an sgRNA, resulting in mutations
in a designated part of the genome (4). We have developed specific sgRNAs for each of the
duplicates of camta1 in zebrafish ( camta1a and camta1b) . The injected embryos were then
screened at several points for any heart deformities and are awaiting genotyping to determine
the efficiency of the mutations.
 To more fully understand the camta1 regulatory network, we are also
To more fully understand the camta1 regulatory network, we are also
interested in locating the regulatory elements of camta1, including its
transcription start site (TSS). 5’ RACE (rapid amplification of cDNA
ends) is used to locate the 5’ end of RNA which can then be aligned
to the genome to discover the location of the TSS.
Results
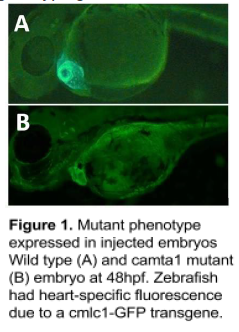
Over the course of the year, we performed multiple microinjections.
Injected embryos exhibited a lethal phenotype of enlarged ventricles
and valve defects (fig. 1) with up to 25% efficiency.
We have also completed the 5’ RACE and will perform sequencing and analysis of our
experimental data at the beginning of the new year.
Discussion
At the beginning of the year, we experienced difficulties with the sgRNA design for camta1b,
which took time to resolve before being able perform microinjections. While we had some
successful injections over the summer, fish with the mutant phenotype died before they could be
genotyped. In order to prove that the fatal phenotype was due to camta1 mutations and not
some other factor, it was necessary to create a homozygous mutant line. To address the issue,
we redesigned our experiment by adding a localization sequence to Cas9, which causes
mutations only to occur in the sperm and eggs.This causes fish to carry a mutation in their
reproductive cells, but not the rest of their body. Injected fish will not express a phenotype but all
of their offspring will. We have injected fish using the new protocol and expect them to reach
maturity in a few months.
The 5’ RACE also presented challenges. This experiment has numerous steps and potential
sources of error. After trying multiple times with inconclusive results, Dr. Hill collaborated with
other researchers to identify a different protocol that may be more effective. This protocol, called
an RLM-RACE, only includes full-length mRNA in the experiment, allowing us to remove noise
caused by degraded or other types of RNA. This protocol was used for our most recent RACE
and we look forward to analyzing the data in the coming year.
Conclusion
The data we have collected strongly indicates a correlation between camta1 mutations and
heart defects. It is, however, inconclusive. By adapting our experimental design, we continue to
progress toward conclusive evidence of the role of camta1 in heart development and CHD.
Sources
1. Hoffman, Julien I.E, and Samuel Kaplan. 2002. “The Incidence of Congenital Heart
Disease.” Journal of the American College of Cardiology 39 (12): 1890–1900.
2. Song, Kunhua, Johannes Backs, John McAnally, Xiaoxia Qi, Robert D. Gerard, James
A. Richardson, Joseph A. Hill, Rhonda Bassel-Duby, and Eric N. Olson. 2006. “The
Transcriptional Coactivator CAMTA2 Stimulates Cardiac Growth by Opposing Class II
Histone Deacetylases.” Cell 125 (3): 453–66.
3. Schwartz, Robert J., and Michael D. Schneider. 2006. “CAMTA in Cardiac Hypertrophy.”
Cell 125 (3): 427–29.
4. Moreno-Mateos, Miguel A., et al. 2015. “CRISPRscan: designing highly efficient sgRNAs
for CRISPR/Cas9 targeting in vivo.” Nat Methods 12 (10): 982-8
