Alec Judd and John Chaston, Plant and Wildlife Sciences
Introduction
It has already been established that gut microbiota affect starvation resistance, fat (TAG) content, and development in D. melanogaster. Previous studies in this lab have suggested that effects of the microbiome on these traits are correlated, but this prediction has not been explicitly tested. We are specifically interested in identifying individual bacterial genes that may mediate microbial effects on all three host traits. One way to identify genes with causal influence on animal traits is MGWA (Meta Genome-Wide Association). To this end we have measured starvation resistance in D. melanogaster individually associated with a panel of 43 bacterial strains, and saw how similar genes in different genomes had similar effects. These became the “interesting” genes, for which we are in the process of proving causation.
Then, we compared our obtained values with previous measures of TAG and development rate conferred by the same strains. Comparing MGWA predictions shows that there are genes that affect each phenotype independently, with some of the less-significant predictions suggesting shared effects. We are currently performing follow-up studies to confirm all of these predictions by mutant analysis.
Methodology
- We prepared fruit flies by dechorionating eggs in order to ensure control over gut microbial composition.
- We mono-associated fruit flies with 43 different strains of bacteria with varying genetic composition.
- With only one type of bacteria in their gut, each group was tested for lifespan under starvation conditions (access to water only)
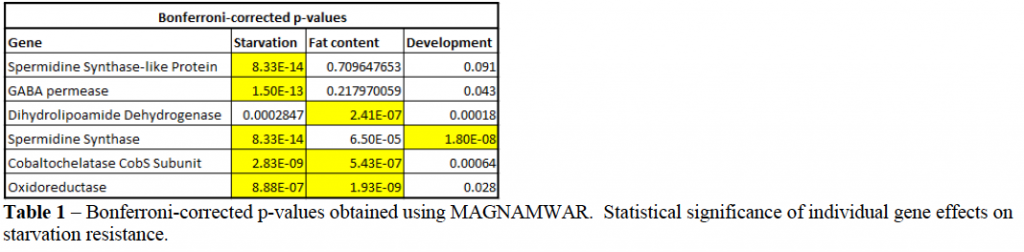
- Results were run through the pipeline “Mono-associated gnotobiotic animal meta-genomewide association R package” (MAGNAMWAR). It provided the predictions listed in Table 1.
Results
Means of starvation time by strain are found in Figure 1.
MAGNAMWAR is designed to calculate the significance of the effect that each bacterial gene has on host phenotype. Of the thousands of genes contained in all the collective bacterial genomes, 861 were found to be significant to p <0.05 (with Bonferroni correction). There is a positive correlation between microbiota effects on triglyceride (TAG) content and starvation resistance, suggesting that host responses to the microbes may be linked. (p-value = 0.01, Rho = 0.45). Similar results are noted when starvation resistance is compared to development time.
Discussion
As seen from the results, we can classify discovered genes into two main categories: genes that affect starvation resistance independently of fat content and development, and those that affect starvation resistance and fat content/development. The evident hypothetical explanation for the effect of genes on starvation resistance is that increased development time leads to increased fat content and fat stores, and there for lengthened starvation time. In addition to proving this hypothesis, it is interesting to note that not all the genes isolated by MAGNAMWAR were significant to all three phenotypes, rather some affect starvation independently, suggesting that the genes involved affect distinct processes in starvation resistance.
Conclusion
All of the results obtained are contributing to the general reservoir of knowledge about the effects that gut microbiota have on their hosts. As we are able to classify genes based on their effects on phenotypes, we can better understand how all genes and proteins work together in biological processes, and extrapolate to orthologous genes and processes in humans. Closer examination of these discovered genes will more easily help us to determine their function, and therefore treat related diseases, either genetic or caused by the environment. Genes that have previously been proven to affect TAG content are currently under experimentation, and mutants of the genes discovered with MAGNAMWAR are being created to prove causation.