Parker Hollingsworth and Stephen Piccolo, Biology
Introduction
Machine learning classification is a type of artificial intelligence that learns from data and makes predictions. There are many different algorithms that can be used to develop predictive models for machine learning. Generally the algorithm looks for patterns in the data and uses those patterns to make predictions on an additional data set. This type of artificial intelligence is being used increasingly in the biomedical community to predict disease diagnosis and prognosis. Although machine learning has shown to provide promising results, it is far from perfect. The accuracy of the predictive model often depends on arbitrary decisions made by researchers. Researchers are often unsure which algorithms or which features of the data to use in the analysis. In this paper, I apply classification algorithms to five datasets and compare the accuracy across 24 different algorithms.
Methods
Data used in this project was downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo). The following datasets were selected for this analysis:
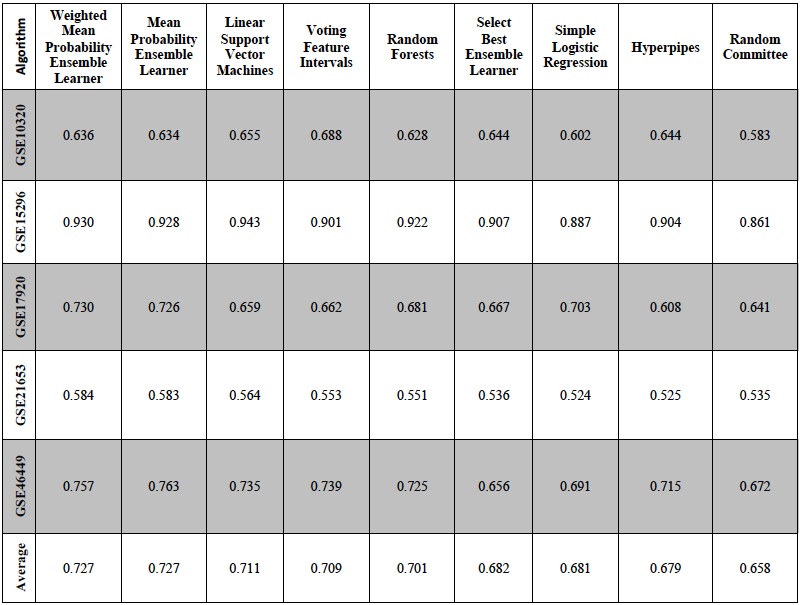
- GSE10320: Predicting Relapse in Favorable Histology Wilms Tumor Using Gene Expression. Data from 53 relapse patients, and 91 non-relapse patients were used.
- GSE15296: Peripheral blood biomarker signatures for acute kidney transplant rejection. Data from 51 acute kidney rejection patients and 24 well-functioning kidney Transplant patients were used.
- GSE17920: Expression data of diagnostic biopsy samples from Hodgkin lymphoma Patients. Data from 38 failed treatment patients and 92 successful treatment patients were used.
- GSE21653: A gene expression signature identifies two prognostic subgroups of basal breast cancer. Data from 83 5-year disease free survival patients and 169 <5-year disease free survival patients were used.
- GSE46449: Expression data from Patients with Bipolar (BP) Disorder and Matched Control Subjects. Data from 49 patients and 39 control subjects were used.
Raw gene expression data were downloaded in the form of .CEL files and normalized using SCAN.UPC (http://www.bioconductor.org/packages/release/bioc/html/SCAN.UPC.html). A tabseparated expression file was created for each dataset. Clinical features were selected manually and added to a separate data file. The class/outcome being predicted was also selected and included in a third file. To perform classification, we used the ML-Flex tool (http://mlflex.sourceforge.net). We applied 24 algorithms to each dataset and evaluated accuracy across 100 randomly selected training and testing data sets. Computing was performed using BYU’s Fulton Supercomputing Lab. The results for each analysis were summarized and put into separate “tidy data” files. This will allow us to easily compare and analyze results across multiple datasets.
Results
We used area under the curve (AUC) scores to compare the predictive accuracy of each algorithm. AUC scores for the nine algorithms that performed best on average across the five datasets are shown below.
Discussion
The results show just how different the predictive accuracy can be depending on the algorithm and type of data used in the analysis. In general, “ensemble” methods performed best; these methods combine evidence across multiple types of data and/or multiple types of algorithms. I am currently extending this research to 95 additional datasets. Our goal is to publish a study in a peer-reviewed journal and make these datasets publicly available to other researchers to perform additional benchmarks. These benchmark datasets will help other researchers to select machine-learning algorithms that are best suited for their own datasets.