Mark Roth and Jonathan Alder, PhD PDBio
Introduction
Telomeres, located at the ends of chromosomes, are repetitive DNA sequences composed of TTAGGG repeated thousands of times. Each time a cell copies its DNA a small amount of telomeric DNA is lost due to the end replication problem. Because of this, telomeres are eventually lost. New telomeres are synthesized by telomerase. Telomerase has both a protein reverse transcriptase component (TERT) and an RNA template (TERC) that it uses to synthesize new telomeres; however, expression is tightly controlled and is not expressed in most cells in our bodies. This limits the number of times that most of our cells can divide. When telomeres get short, they trigger a DNA damage response that leads to senescence or apoptosis. Telomeres are longest when we are born and they shorten as we age. While the role of short telomeres in cellular apoptosis and senescence is well established, less is known about the gene expression changes that occur when telomeres shorten. In some pathogenic yeast, it has been shown that expression of genes located next to the telomere changes when the telomeres become short, due to epigenetic changes in the DNA1. This phenomenon has been termed subtelomeric silencing. Genes in the area close to the telomeres are suppressed by the telomeres when they are long, but when they are shortened the genes are expressed in greater frequencies. Here we will test if subtelomeric silencing occurs in human cells and examine if telomere shortening could underlie age-related changes in gene expression. This phenomenon could also play a role in certain age-associated diseases.
Methodology
Most cell lines commonly used in cellular biology are immortalized. This means that they have overcome the end replication issue. The most common way that this occurs is through activating telomerase. These are commonly cancer cells as well, meaning that they have oncogene mutations and grow and divide at a rapid pace. We wanted to be able to study cells as they age, requiring us to create a conditional cell line that was telomerase negative. However, the oncogene mutations are beneficial because they allow us to study the effects of a lifetime of aging in a couple of months. Creating a conditional cell line was more difficult than originally planned. We started with several attempts to insert lox-P sites on both sides of TERC and create a cell line that stably expressed CreERT2. This failed because we were unable to homozygously introduce these Lox-P sites on several occasions.
We overcame this by coming up with an alternative method. Using CRISPR/Cas9 – mediated genome editing we created a TERT knockout (KO) cell line using a sgRNA that guides the Cas9 protein to the catalytic site of the TERT gene. The sgRNA was transduced into a line of K562 cells that already stably expressed Cas9. The viral DNA that contained the sgRNA also encoded for puromyacin resistance. These cells were then exposed to puromyacin (2mg/mL). This cell line was then individually cloned and grown out. We confirmed that telomerase was inactive using a TRAP assay and sequenced the catalytic site of the TERT gene to confirm that TERT was inactive due to a frameshift mutation. This cell line was then allowed to divide and replicate until its proliferation ceased. This took roughly 70 days from the time of sgRNA addition and selection (figure 1). Unfortunately due to the setbacks already described, as well as the characterization of the cell line taking longer than anticipated, this was as far as we were able to get to date on the project that we originally proposed. We are continuing work with this now-characterized cell line.
Results and Discussion
Although we have not completed the project at this time, we have created a proof of concept. We took a cell line that was telomerase positive and immortalized, and we inhibited telomerase activity, confirmed that it was telomerase negative, and showed that after a specific length of time the cell line would senesce. We kept meticulous cell counts of each split and showed that the cells stop dividing around 70 days after the introduction of the sgRNA (figure 1).
Conclusion
The ability to create and characterize telomerase-negative cell lines will be vital for our research as we continue to look into our hypothesis that subtelomeric silencing occurs in humans. The next step is to create a new telomerase KO cell line and use the timeline that we established in this project to collect RNA for RNAseq and analysis. We will collect genomic DNA at several time points to clearly show (via Southern Blot) that the telomeres are shrinking. This KO cell line also will allow us to study and find other genes that may “rescue” a cell line from senescence.
Figure 1
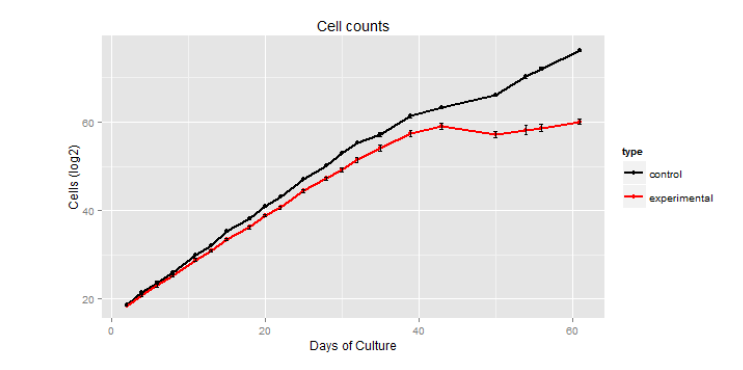
This shows that the experimental cells stopped dividing (red), while the control cells continued (black). It also shows that this change occurred around day 40 of counting cells, which was around day 70 of cell growth after sgRNA introduction.
1 Irene Castaño, Shih-Jung Pan1, Margaret Zupancic, Christophe Hennequin, Bernard Dujon and Brendan P. Cormack. (2005) Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Molecular Microbiology 55: 1247-1258.
