Caj Johansson and Byron Adams, Biology Department
Introduction
The current belief of the majority of glaciologists is that during the last glacial maximum (LGM), 17,000-22000 years ago, the entire Antarctic continent was completely covered in ice (Convey, 2008). This would mean that all terrestrial organisms would have died out during this time, essentially leaving Antarctica without life. If this hypothesis is true then all terrestrial organisms found currently in Antarctica arrived after the LGM (the past 12-18,000 years), a very short time frame for speciation to occur. Tardigrades are microscopic terrestrial animals that are most closely related to Arthropods and Nematodes (Edgecombe, 2011 but see also Longhorn, 2007). They are able to survive the cold, dry conditions present in Antarctica along with only a handful of other organisms. Tardigrades are extremophiles, capable of surviving extreme temperatures, pressures, levels of radiation, dryness, and chemical toxins. Because of this extraordinary survivability, tardigrades are ideal candidates for having survived the harsh conditions present during the LGM. My hypothesis is that tardigrades did in fact survive the LGM and were present on the Antarctic continent long before this time.  This is an important hypothesis because if it is true, then it would mean not only that the majority of Antarctic Glaciologists are wrong about the extent of the Antarctic ice sheets during the LGM, but also that small isolated pockets of suitable soil habitat remained where the organisms were able to survive. With sufficient proof that these areas of exposed soil existed, current ideas about the glacial history of Antarctica would have to be rewritten.
This is an important hypothesis because if it is true, then it would mean not only that the majority of Antarctic Glaciologists are wrong about the extent of the Antarctic ice sheets during the LGM, but also that small isolated pockets of suitable soil habitat remained where the organisms were able to survive. With sufficient proof that these areas of exposed soil existed, current ideas about the glacial history of Antarctica would have to be rewritten.
Methodology
Every year Dr. Adams travels to Antarctica to collect soil samples. These samples are then shipped to our lab and under a microscope we examine them and pull out the individual tardigrades. Then the DNA of the tardigrades is then extracted using a worm lysis buffer protocol. Next PCR is performed to amplify the ribosomal 18s and 28s regions of DNA and also mitochondrial DNA from the COI gene. Then the PCR product is cleaned using a Gene Clean II kit and then a Big Dye cycle sequencing reaction is performed using the same primers that were used in the PCR reaction. The samples are then sent off to the BYU sequencing center to be sequenced. The results are then downloaded and edited using the program Geneious (version 7.0.4, Biomatters; http://www.geneious.com/). The different DNA sequences will then be aligned and the differences between them will be analyzed via phylogenetic analysis [Andrew Thompson, a Biology graduate student, analyzed the sequence data using TNT (parsimony analysis software) and Mr. Bayes, which depends on Bayesian statistics. Prior to these analyses, sequences were aligned use MAFFT].
Results
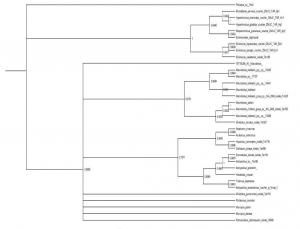
While we have assembled all of the DNA sequence data that is necessary, we are still in the process of analyzing these sequences and compiling them into credible, publishable phylogenetic trees. Preliminary trees have been produced, (see figure 2) but they are preliminary and not to the necessary quality yet to be published. These preliminary trees seem to support our alternative hypothesis.Discussion
Autapomorphies among homologous nucleotides are evidence of lineage independence (speciation). The more autapomorphies that are present in the DNA of two organisms, the longer that they have been evolving independently. My null hypothesis is that tardigrades did not survive the Pleistocene (colonized Antarctica after the LGM). Direct evidence for this would be if all the tardigrades currently in Antarctica are also found elsewhere in the world, and that populations recently (last 12,000 years) established in Antarctica via dispersal. Additional support for this hypothesis would come from evidence showing that there is no significant difference in genetic diversity among the populations at higher elevations, which could have served as refugia during the Pleistocene, versus lower elevations near coastal regions that have most recently undergone deglaciation. My alternative hypothesis is that tardigrades were present in Antarctica before the LGM and that they were able to survive it by taking refuge in small areas of suitable habitat that remained ice free throughout the LGM. These areas would have been present at very high elevations where the ice would have not covered them. Evidence supporting my alternative hypothesis must demonstrate higher levels of genetic diversity at the high elevations than the low elevations. This would support the idea that tardigrades survived the LGM and then subsequently colonized habitats at lower elevations as the ice receded.
Conclusion
Although our hypothesis remains a bit ambiguous for now we have learned some valuable information from our preliminary results. We have learned that the 18s gene region is highly conserved among individuals and thus is best for analyzing differences between different species or different isolated populations. The COI gene region however is very variable and can be used to characterize genetic differences between individuals of the same population. This will be invaluable in subsequent tardigrade phylogenetic studies. We will complete the analysis of our entire data set by the end of the year and publish our results in the journal Polar Biology or another similar journal this coming spring.
Scholarly Sources
Convey, P., J. A. E. Gibson, C. D. Hillenbrand, D. A. Hodgson, P. J. A. Pugh, J. L. Smellie, and M. I. Stevens. 2008. Antarctic terrestrial life – challenging the history of the frozen continent? Biological Reviews 83:103-117.
Edgecombe, G. D., G. Giribet, C. W. Dunn, A. Hejnol, R. M. Kristensen, R. C. Neves, G. W. Rouse, K. Worsaae, and M. V. Sørensen. 2011. Higher-level metazoan relationships: recent progress and remaining questions. Org. Divers. Evol. 11:151-172.
Longhorn, S. J., P. G. Foster, and A. P. Vogler. 2007. The nematode-arthropod clade revisited: phylogenomic analyses from ribosomal protein genes misled by shared evolutionary biases. Cladistics 23:130-144.