Jeffrey Schachterle and David Erickson, Microbiology and Molecular Biology
Introduction
The bacteria Yersinia pestis is the causative agent of bubonic plague, and its ability to form biofilm in fleas is essential for plague transmission by fleas [1]. Y. pestis recently evolved from Y. pseudotuberculosis (Y. pstb), and the two have nearly identical genomes. Y. pestis forms a biofilm in fleas that is capable of blocking the flea’s feeding. These biofilms are made up of a community of bacteria adhering to a polysaccharide matrix that is made by the hmsHFRS gene products. Y. pstb is capable of infecting fleas, but will not block the flea’s feeding [1]. Since the hmsHFRS genes themselves are identical, this suggests that Y. pstb has regulatory elements that function differently from those of Y. pestis [2]. Although the hms genes in these two bacteria likely differ in regulation only, there may be other exopolysaccharide producing genes that may account for different biofilm properties. Understanding the manner in which biofilm is regulated in Y. pstb will aid in understanding the regulation of biofilm in Y. pestis and the manner in which they have evolved the ability to be transmitted through fleas.
The Erickson lab has previously shown that the BarA/UvrY two-component system indirectly regulates biofilm production in Y. pstb through the csr system. Proteomic analysis comparing uvrY and csrB mutants to wild-type identified a myriad of candidate CsrA target genes. In order to reduce the number of candidate genes to those regulating biofilm production, I have conducted a transposon mutagenesis in the high biofilm forming csrB mutant to identify suppressor mutations. The findings of the suppressor mutation screen suggest that O-antigen synthesis of the lipopolysaccharide may be important for biofilm formation. I have also tested the global regulator, Hns, for a role in biofilm formation. These findings suggest that while Hns appears to play a role in biofilm formation, it is likely through exopolysaccharides other than those produced by the hms operon.
Materials and Methods
Transposon mutants were generated via tri-parental matings that utilized a B083 E. coli strain containing pJG285 as the donor, B001 E. coli as a helper strain, and the Y. pstb csrB mutant as the recipient. These matings were plated onto nutrient plates containing Congo Red dye as well as antibiotics to select against the donor and helper E. coli strains as well as the Y. pstb csrB cells that had not been mutagenized. The cells were plated so that about 200 colonies grew per plate. From these plates, the colonies that had taken up the least amount of the Congo Red dye were selected. In this manner, between 4,000 and 5,000 colonies were screened. For each selected colony, the region flanking the transposon was amplified using arbitrary PCR primers. This amplified region was then sequenced to identify the transposon insertion sites. This was accomplished by utilizing the BLASTn available from www.ncbi.nlm.nih.gov.
Testing of the role of Hns on biofilm formation was conducted by cloning the full length hns gene into the pJET1.2 plasmid behind a constitutive promoter. A dominant-negative form of hns was also cloned into the same plasmid background. These plasmids were transformed into Y. pstb strains including wild-type, uvrY mutant, and csrB mutant. This was accomplished via electroporation. These strains were then assessed for biofilm formation quantitatively by crystal violet staining following growth in liquid media, or qualitatively by Congo Red binding when grown on agar petri plates.
Results
39 mutants were sequenced. Interestingly, no mutations in any of the hmsHFRS operon were found. However, 5 mutations to genes in an O-antigen biosynthesis operon were found in the suppressor mutation screen.
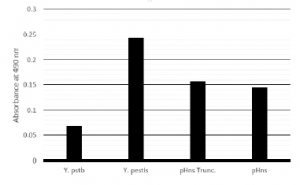
Results of biofilm assays have been varied regarding the role of hns on biofilm formation. When grown in liquid culture and quantified, expression of the dominant negative form of hns leads to increased biofilm, whereas, expression of the full length hns does not significantly increase biofilm formation as shown in Figure 1. When grown on agar plates containing Congo Red dye, neither expression of the dominant-negative hns nor the full length hns alters morphology or pigmentation of the cells.
Discussion and Conclusions
The finding that 12.8 percent of mutations that were able to suppress the high biofilm formation phenotype of the csrB mutation were in key O-antigen biosynthesis genes suggests that O-antigen may play a significant role in Y. pstb biofilms. Because the biofilm of Y. pestis is almost exclusively poly-N-acetyl glucosamine produced by the HmsHFRS proteins, this may suggest that the differences between Y. pstb and Y. pestis biofilm formation may be more than regulation of the hmsHFRS genes. The importance of O-antigen in Y. pstb, along with the fact that Y. pestis does not produce O-antigen, suggests that there are important structural differences in these biofilms as well as regulatory differences.
Because hns appears to be regulated by the csr system and hns has an impact on biofilm, this suggests that at least one of the mechanisms by which the csr system regulates biofilm production in Y. pstb is by regulating the global regulator hns. Combined with the finding that there are likely other exopolysaccharides that are significant to Y. pstb biofilm formation, this suggests that there are complex regulatory and structural variations in these biofilms. This additional complexity in Y. pstb compared to Y. pestis biofilms is likely due to selective pressures applied by the wider variety of environmental niches occupied by Y. pstb.
References
1. Erickson DE, et al. 2006. Serotype Differences and Lack of Biofilm Formation Characterize Yersinia pseudotuberculosis Infection of the Xenopsylla cheopis Flea Vector of Yersinia pestis. J Bacteriol. 188(3): 1113–1119.
2. Sun YC, et al. 2011. Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One 6:e19267.
Figure 1: Biofilm assay based on Crystal violet staining of cells adherent to culture tube