Matthew Moulton with Dr. Michael Whiting, Department of Biology
Introduction
Mitochondria are unique organelles that contain their own genomes. Metazoan mitochondrial genomes typically encode 37 genes (13 protein-coding, 22 transfer RNA, and 2 ribosomal RNA genes) that are used for cellular respiration. Mitochondrial DNA (mtDNA) sequence data have proven to be an indispensable tool for using molecular systematics to infer relationships among species and the number of complete mitochondrial genome sequences made available for researchers has increased rapidly over the past decade. The mitochondria inside cells rely heavily on genes encoded in the nucleus for some of its functions such as genome replication, but interactions between nuclear DNA and mitochondrial DNA are not well understood One such interaction that has been shown to occur, but remains poorly understood is a mechanism that causes copies of the mitochondrial genes to be incorporated into the nuclear genome. These copies become non-functional pseudogenes and are known as nuclear mitochondrial pseudogenes, or numts. Numts have been found in many organisms and inadvertently sequencing a numt instead of the mitochondrial ortholog can cause incorrect phylogenetic inference. Coamplification of numts has been problematic for some phylogenetic studies and has been a major cause of incorrect species identification and overestimating the number of species using DNA barcoding methodologies. Understanding the extent to which numts are present in organisms and the evolution of numts over time is critical for correct phylogenetic inference; my research focuses on improving this understanding. Numts have been found in many insect lineages and are especially prevalent within Orthoptera (grasshoppers, katydids, crickets, and wetas). Numts from orthopteran lineages have also proven to be fairly easy to amplify and clone in previous studies, making them ideal taxa to use for my research.
Objectives
I set out to test the following hypotheses in my research: (1) To what extent are numts of three mitochondrial genes (Cytochrome Oxidase Subunit I and II, and NADH Dehydrogenase Subunit V) present within Orthoptera? (2) What is the pattern of evolution of numts within and among individuals? (3) Do numts of the three individual genes show congruent patterns of evolution?
Materials and Methods
I selected thirty-six orthopteran taxa (representing 34 families from both suborders of Orthoptera, Ensifera and Caelifera) for use in this project. I extracted the DNA from these individuals using the QIAGEN DNeasy kit. I amplified the three gene regions (COI, COII, and ND5) via PCR using Elongase® Enzyme mix and forward and reverse primers sets. The polymerase in this enzyme mix is designed to have a very low error rate (0.0015%/bp). The PCR product were cloned using the TOPO TA Cloning® Kit (Invitrogen Corporation). Fifty clones from each reaction were picked and DNA from each clone was amplified via PCR using the same enzymes, and protocol used for the original amplification above. PCR products were cleaned using USB® PrepEase® PCR purification 96-well plate kit and sequenced using BigDye® chain terminating chemistry. Each sequencing reaction was filter-cleaned and fractionated on an automated sequencing machine (ABI 3730xl) in the BYU Sequencing Center. Sequence data was assembled and proofed in Sequencher 4.9 (Gene Codes Corporation). I have obtained the orthologous sequences for all taxa and all genes from a member of the Whiting lab who is sequencing the entire mitochondrial genomes for all the taxa in my project. A comparison of each clone sequence with its corresponding ortholog was made in Sequencher in order to identify putative orthologs, heteroplasmy, and numts by analyzing and quantifying in-frame stop codons, base composition, indels, and point mutations. Although phylogenetic analyses have not yet been performed, an alignment using MUSCLE will be made and phylogenies will be reconstructed using the aligned sequences in both a parsimony and a Bayesian framework using TNT and MrBayes, respectively, in order to investigate patterns of evolution.
Results and Discussion
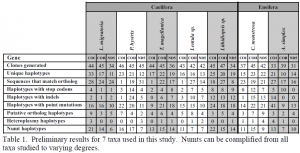
Analyses have shown that numts of COI are present in several lineages of Orthoptera. I currently have a manuscript in press in the journal Molecular Ecology Resources detailing this discovery. To date, I have collected sequence data of 88/108 genes from 35/36 taxa. This represents about 81% completion of all the data collection. I am continuing to work in the Whiting lab post graduation to complete this project. I anticipate completing all data generation by July 20th and will have the phylogenetic analyses run by July 31st. I will then work with my collaborators to compile the findings from this study into a manuscript to be submitted to a peer-reviewed journal by the end of 2010. The preliminary results of my findings confirm that numts of COI, COII, and ND5 are all present in all lineages tested to varying degrees. Table 1 shows the preliminary results for some taxa that have data from all three genes collected. Although, we have yet to reconstruct phylogenies of these numts, it appears that numts have occurred many times throughout the evolutionary history of these taxa. I anticipate that the findings from this study will help solidify molecular techniques for identifying and quantifying numts as well as lay a foundation for future numt studies in other organisms.