Mac Martin with Dr. Byron Adams, Department of Biology
Glaciologists have suggested that because of the extent of the ice sheets during the last glacial maximum (LGM) in Antarctica no terrestrial species could have survived these harsh conditions. However, substantial evidence suggests that some invertebrate species did indeed survive the LGM. We hypothesize that S. lindsayae survived the LGM in Antarctica through hypothetical refugia, areas that remained ice-free during the LGM. These are coastal areas characterized by high elevation, which would have remained ice-free during the LGM and would not have been inundated under episodes of elevated sea levels.
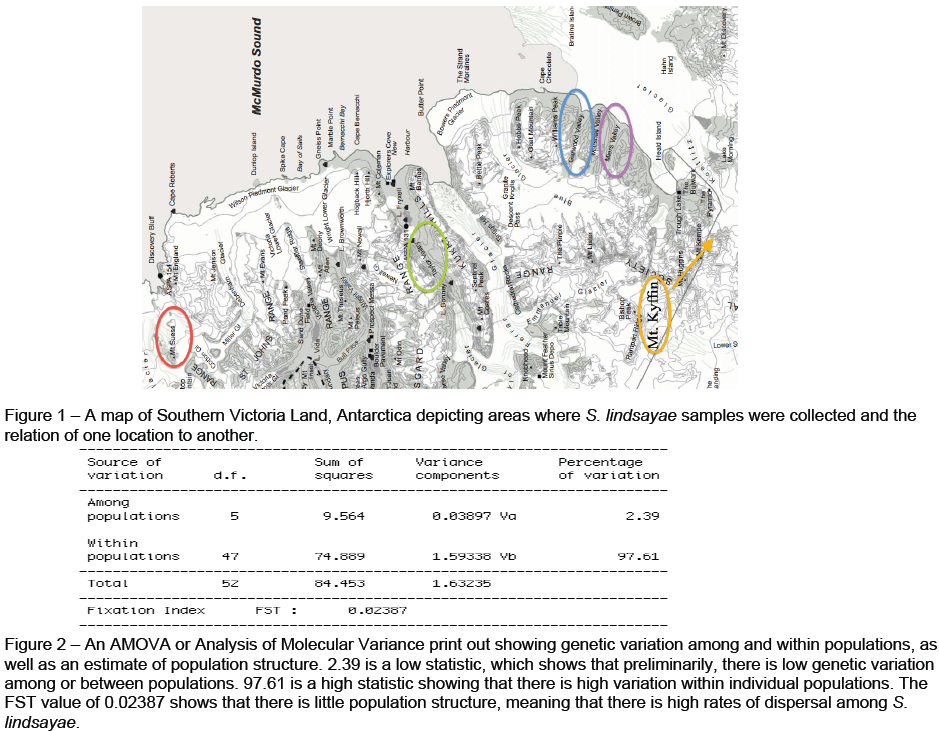
In the first stages of this research project we hypothesized that the location of Mt. Seuss in Antarctica served as a refugium for S. lindsayae because of its elevation and costal location. We determined that if we could show greater genetic variation of S. lindsayae from Mt. Seuss to other neighboring locations, which would have been completely inundated by ice during the LGM, this would support our refugia hypothesis. We began to collect nematode samples from Mt. Seuss and from the hypothetical sink locations such as Taylor Valley, Garwood Valley, Miers Valley, and Mt. Kyffin.
After extracting the DNA from individual nematodes, we subsequently performed Polymerase Chain Reaction (PCR) to amplify a 195 bp fragment of mtDNA from each nematode. After checking for amplification of the fragment through gel electrophoresis, we used the TOPO TA cloning kit. Because the size of the target fragment was so small, we cloned each individual so that we would have cleaner target DNA before we tried to sequence it. We performed PCR on the clones using M13 primers, designed for the pCR 2.1 vector, and subsequently cycle sequenced the amplified product. We trimmed off the vector and assembled contig sequences using Sequencher and Codon Code Aligner. We then had to align and edit sequences with ClustalX and MacClade. Next, the sequences were taken and a neighbor joining phylogenetic tree was constructed with PAUP. Population genetics analyses were performed using Arlequin and Structure to show how one population compared to another in terms of genetic variation.
Although this project is not yet completed, we found high genetic variation among all S. lindsayae sampled. There were 34 unique haplotypes among the different populations. Many of these haplotypes were due to polymorphisms or variation within an individual nematode. Results of population genetics analyses show low genetic variation among or between populations and high variation within a population. Preliminary data show significant levels of genetic variation but low population structure and high dispersal rates among populations of S. lindsayae. We expected to see high population structure and high genetic variation between populations such as between hypothetical source and sink populations. Failure to detect this signal, is likely due to insufficient sample sizes of individuals from Mt. Seuss, our hypothetical refugium.
Even though our preliminary data is not conclusive evidence in support of the hypothesis that S. lindsayae predated the LGM through hypothetical refugia, the fact that this species is endemic to continental Antarctica suggests strong support of the refugia hypothesis. Also, because we have already detected high genetic variation among S. lindsayae individuals, we show that it is quite possible that conclusive evidence of a refugia hypothesis is near. Additional sampling of previously collected material is already underway. By greatly increasing our sample size of Mt. Seuss we will be able to detect variation among or between population groups, which we hypothesize will be in support of the refugia hypothesis. Our ultimate goal is to look for concerted patterns of distribution of numerous biota in order to get a better idea of how biotic communities have responded to historical patterns of environmental change.
I was able to present my research and preliminary data at the Society of Nematologists 49th annual meeting in Boise, ID on July 11-14, 2010 and at the McMurdo LTER Science meeting in Fort Collins, CO on August 27-28, 2010. I was able to orally present my research in Boise and I made a poster with a summary of my research for the meeting at Fort Collins. I anticipate publishing an article summarizing my final results in the Journal of Nematology.
I would like to thank ORCA for funding my research, Dr. Byron Adams and the BYU Nematode Evolution Lab for all their help along the way, and Diana H. Wall for her input and help in review of abstracts.