Jared Lambert and Dr. Brian Poole, Life Sciences
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease which commences from the immune system producing antibodies which target the body’s own tissues and cells. There are various factors thought to be involved in the development of SLE. In this study I assessed how the splicing variants of the interferon regulatory factor (IRF5) gene are affected by a single nucleotide polymorphism (SNP) associated with risk for systemic lupus erythematosus (SLE). Through this assessment I hoped to more fully determine the mechanisms through which this risk allele affects the development of SLE.
IRF5 is a cytoplasmic transcription factor gene responsible for interferon production. Polymorphisms of IRF5 are genetically associated with SLE (2). A trait of SLE patients is the presence of elevated levels of interferon (proteins that are invaluable in the immune systems response to viral infection) (3). Previous research has shown that the risk of SLE is strongly associated with the single-nucleotide polymorphism (SNP) rs2004640 “T” allele in IRF5. This has been confirmed multiple times across many ethnicities (4). This specific allele creates a novel splice acceptor site in the RNA. My research focused on obtaining data on how the systemic lupus IRF5 risk haplotype affects the cells of the immune system by assessing its splicing variants through PCR and DNA sequencing.
Methods
Through TaqMan quantitative polymerase chain reaction (PCR) the lab was successful in genotyping over 100 volunteers. After genotyping it was possible to create eight matched sample sets based on gender and ethnicity matched homozygous risk and homozygous protective samples at the SNP rs2004640. Following, was the assessment of the splicing variants of the IRF5 risk haplotype through PCR and DNA sequencing. For mRNA expression level studies of the first exon of IRF5, quantitative PCR was used to determine relative abundance of exons 1A, 1C, and 1D in risk versus protective cells. Statistical analysis was done using the Wilcoxon Signed Rank Sum test for paired samples.
Results
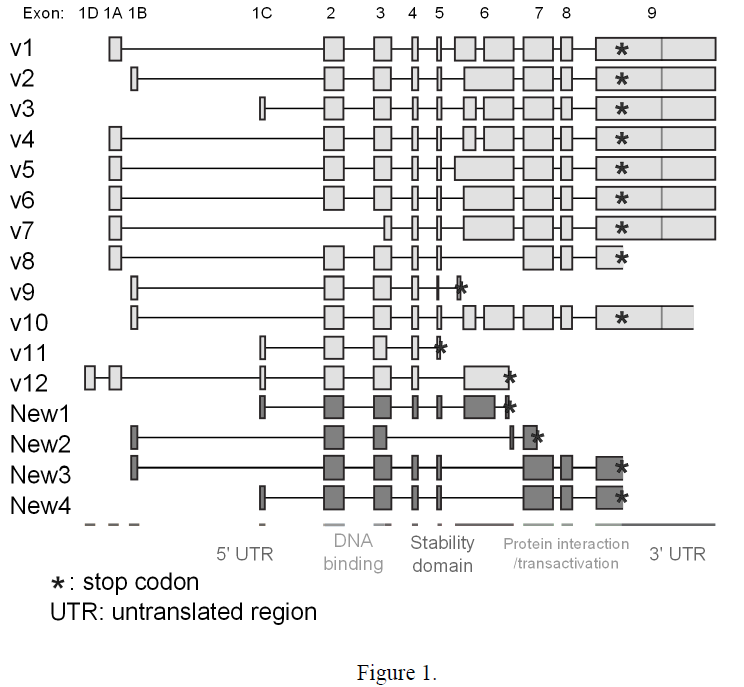
The goal of the experiment has been to determine which variants may contribute to the aberrant interferon response observed with SLE. Through previous research IRF5 has been found to be spliced at least 12 different ways. Exons 1A, 1B, 1C, and 1D may be alternatively spliced as exon 1 in the processed mRNA. The presence of the SNP produces a novel alternative splice site which leads to the presence of exon 1B exclusively in cells with the risk allele (1). I analyzed the remaining exons in order to see differences in exon 1 usage between those with the risk haplotype and those with the protective haplotype. Testing of the expression levels of exon 1A, 1B, 1C, and 1D of IRF5 through PCR, it was found that exons 1C and 1D are under-expressed in risk cells.
Through the sequencing of exons 1 to 7, identification of five novel splicing variants of IRF5 have been discovered (Figure 1). New 1, 2 variants were found to be missing the stability
domain and as a result were able to bind DNA but lacked the ability to initiate transcription. The new 3 and 4 variants were found to have part of stability domain missing. New 5 variant was able to bind to DNA but could not trans-activate. These novel variants have not been found to be more or less abundant in the risk cells.
Conclusions
We should change how we view the splicing of this gene as there may not be a set number of variants. There are likely dozens of ways to splice IRF5. The function of the risk allele may be in a shift of balance in the splicing of exon 1. In protective cells there is more mRNA using exons1C and 1D than risk cells. 1A is not altered by the presence of the risk allele (data not shown). Exon 1B is only in risk. Thus, 1C and 1D variants may perform a protective role. Likewise, the presence of exon 1B variants may perform a risk-conferring role.
This establishes a risk splicing profile: risk allele-containing cells exclusively contain exon 1B, which in turn have less 1C and 1D.
Through these findings it is clear that there is still much to be learned and discovered about the IRF5 gene. There have been factors discovered through this research but it is apparent that there are still factors yet to be discovered.
References
- Robert R Graham, Sergey V Kozyrev, Emily C Baechler, MV Prasad LingaReddy, et al. “A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus.” Nat Genet. 2006 May;38(5):550-5. Epub, 2006 Apr 16.
- Graham RR, KyogokuC, Sigurdsson S, VlasovaIA, Davies LR, BaechlerEC, et al. “Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypesfor human lupus.” Proc NatlAcadSciU S A 2007;104(16):6758-63.
- KozyrevSV, Alarcon-RiquelmeME. “The genetics and biology of Irf5-mediated signaling in lupus.” Autoimmunity 2007;40(8):591-601.
- ManclME, HuG, Sangster-GuityN, OlshalskySL, Hoops K, Fitzgerald-BocarslyP, et al. “Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. Multiple isoforms with distinct cell type-specific expression, localization, regulation, and function.” J Bio Chem2005;280(22):21078-90
