Dohyup Kim and Dr. Keith Crandall, Biology Department
Introduction
The order Decapoda represents a species-rich and economically important group of crustaceans. Among decapods, lobsters play a huge role of monetary significance, bringing in billions of dollars each year to world fisheries. Understanding the genetic relationships among commercially important lobsters will aid in both fishery and conservation efforts. The infraorder Glypheidea was thought to be extinct until 1975, when the species Neoglyphea inopinata was discovered in the Philippines (Forest et al, 1976). Today, this species is considered a “living fossil” and examining the phylogenetic placement of this species in relation to other groups will shed light on the evolution and origins of lobster-like decapods. Although the infraorders Achelata, Asctacidea, Glypheidea, and Polychelida are considered lobster-like decapods, they each possess a suite of morphological characters unique to members of their own group. The vast degree of morphological disparity among these infraorders will allow us to study the evolution of particular traits. For example, claw shape can vary tremendously from the massively bulbous and elongated (and tasty) claws seen within the Astacidea to the complete absence of claws as seen in the Achelata. Other groups such as the polychelids and glypheids show alternative forms. Another morphological trait that can be mapped upon a well-resolved phylogeny is the reduction or loss of eyes. Members of the polychelids and astacideans contain members that are blind and associated with the deep sea. Tracing this character throughout the evolution of lobster-like decapods will allow us to correlate the loss or reduction of a functional eye with habitat. It will also be interesting to examine whether lobster-like decapods originated in the deep sea and colonized shallow waters or vice versa.
Project Goals
The goal of this project is to examine the phylogenetic relationships of lobsters within the Decapoda, compare our tree against existing hypotheses of lobster evolution, and study the origins of key innovations and morphological traits using molecular data. The resulting phylogeny will provide us with a testable hypothesis. We will make comparisons between our tree and current morphological and molecular hypotheses. We will test the monophyly of each infraorder and explore the relationships among major lineages. The high degree of morphological plasticity within lobsters will allow us to examine the origins and/or loss of unique adaptations such as clawed pereopods (legs) and the modification of light-sensing systems.
Materials and Methods
* Targeted gene regions amplified using the polymerase chain reaction (PCR).
* Three nuclear loci (18S, 28S, H3) and 1 mitochondrial loci (16S) selected examined.
* PCR products purified using filters (PrepEase PCR purification 96-well Plate Kit, USB Corporation) and
sequenced with the ABI Big-Dye Terminator Mix (Applied Biosystems, Foster City, CA).
* Sequencing products run (forward and reverse) on an ABI 3730xl DNA Analyzer 96-capillary automated
sequencer.
* Sequences assembled and cleaned using Sequencher 4.8.
* Alignments preformed in MAFFT or MUSCLE. GBlocks v0.91b (Castresana, 2000) used to omit highly
divergent regions.
* Models of evolution determined in MODELTEST or jMODELTEST.
* Maximum likelihood (ML) and Bayesian Inference Analyses (BAY) conducted on a partitioned dataset
* Confidence in the topology estimated with bootstrap support values and posterior probabilities.
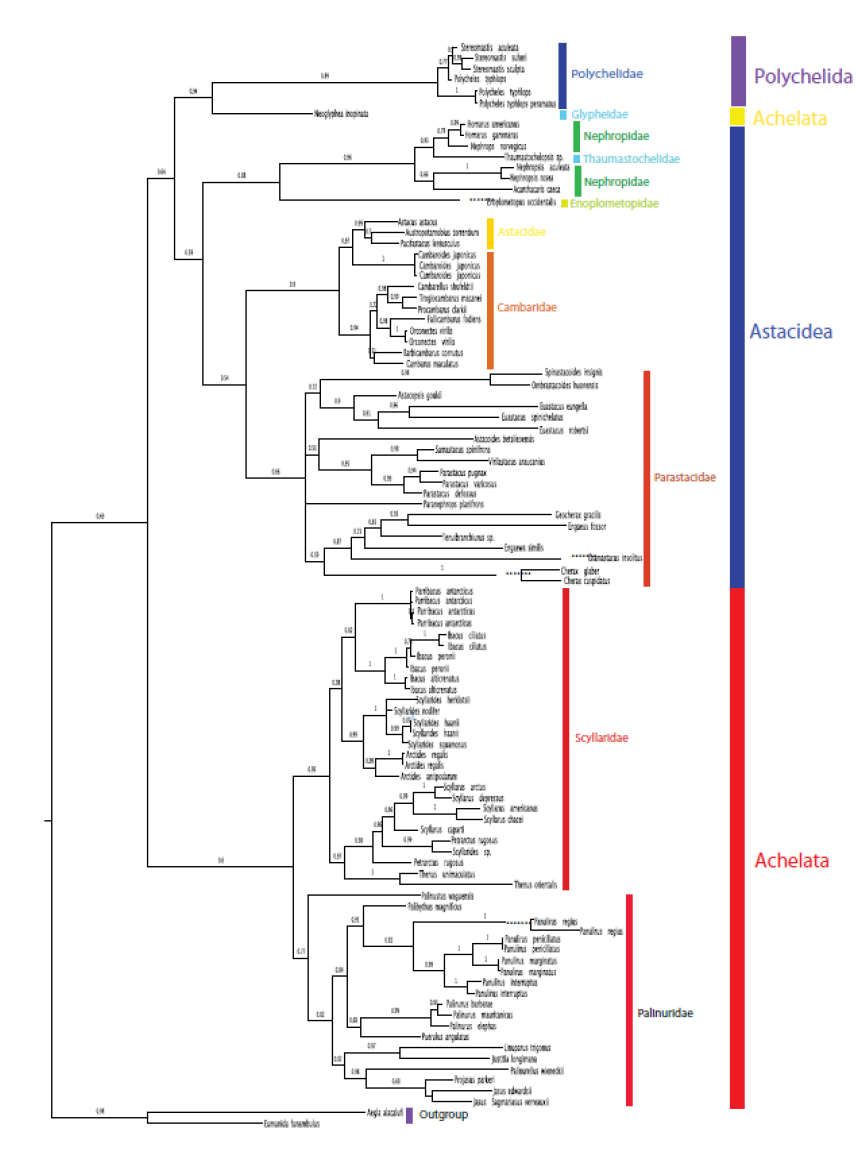
Results
The result of the analysis is represented in a following phylogenetic tree.