Kevin Hiatt and Dr. Michael Whiting, Department of Biology
Introduction
The insect order Orthoptera (grasshoppers, katydids, and crickets) is the most diverse member of the group of polyneopteran insect orders, with more than 23,000 described species (Kevan 1982). Previous studies have recovered the monophyly of the two orthopteran suborders Caelifera (grasshoppers) and Ensifera (crickets and katydids); however, the family-level phylogeny of the order remains largely unresolved. Flook et al. (1995) sequenced the first entire orthopteran mitochondrial genome (mtgenome), and 25 complete mtgenomes are currently available on GenBank. These available mtgenomes only represent two caeliferan and four ensiferan families. Flook et al. (1995) also discovered a tRNA rearrangement (a reversal of the ancestral order tRNALys-tRNAAsp) that Fenn et al. (2008) later proposed to be a synapomorphy for Caelifera.
Objectives
The purpose of this study was to generate a more thorough sampling of orthopteran mtgenomes and to reconstruct a phylogeny of Orthoptera in order to address the following questions: Are the suborders Ensifera and Caelifera monophyletic? Are the diverse superfamilies Acridoidea and Tettigonioidea monophyletic? Is there a pattern of phylogenetic signal present in tRNA rearrangements in Orthoptera? We discuss these questions in light of our findings and also evaluate the utility of mtgenomes in phylogenetic analysis.
Materials and Methods
Taxon sampling
We sequenced entire mtgenomes for six orthopterans from six previously unrepresented families: Ellipes minuta (Tridactylidae), Physemacris variolosa (Pneumoridae), Lentula sp. (Lentulidae), Prionotropis hystrix (Pamphagidae), Lithidiopsis sp. (Lithidiidae), and Xyleus modestus (Romaleidae). We extracted DNA for each of these taxa from specimens available in the Insect Genomics Collection at Brigham Young University. We selected six polyneopteran taxa as outgroups for our phylogenetic analyses.
DNA extraction, PCR and Sequencing
We extracted DNA using the QIAGEN DNeasy kit. We designed Orthoptera-specific primers and used them to amplify the DNA using Elongase® Enzyme mix. We sequenced the PCR products using BigDye® chain-terminating chemistry and an ABI 3770xl automated sequencer (BYU Sequencing Center). We then designed species-specific primers to amplify and sequence the remainder of each mtgenome. We performed nested PCR on long PCR template (greater than 4kb) to avoid coamplification of nuclear mitochondrial pseudogenes.
Sequence analysis and phylogenetic methods
We proofed sequence files using Sequencher 4.9. We uploaded and annotated complete mtgnome sequences in MOSAS (http://mosas.byu.edu). We translated protein-coding genes into amino acid sequences and aligned them using MUSCLE. We then back-translated the aligned amino acid sequences into DNA sequences. We also aligned ribosomal RNA sequences using MUSCLE. We aligned transfer RNA sequences based on secondary structure using INFERNAL. We reconstructed the phylogeny of Orthoptera based on amino acid sequences under a parsimony (TNT) framework and based on nucleotide sequences under a Bayesian (MrBayes) framework.
Results
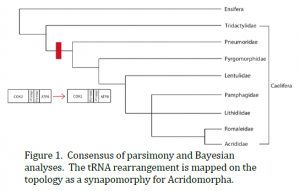
We sequenced all six mtgenomes in their entirety, including the control region. Our parsimony analysis recovered one most parsimonious tree. This topology recovered Ensifera as paraphyletic, while supporting the monophyly of Caelifera and Acrididae. Our Bayesian analysis recovered a single topology that supported the monophyly of Ensifera and Caelifera, but recovered Acrididae as paraphyletic because X. modestus nested within Acrididae. We combined these two topologies by strict consensus to create a family-level phylogeny of Caelifera (Figure 1). The topologies disagreed on the relationship between the families Pamphagidae and Lithidiidae and the sister taxa Romaleidae and Acrididae, so this node was collapsed. Because we have not yet finished our mtgenome sampling for Ensifera, we did not include our additional ensiferan taxa in these analyses and therefore did not include Ensifera in this family-level phylogeny consensus. We also found that E. minuta, which grouped as the most basal caeliferan taxon, did not have the tRNA rearrangement thought to be conserved across Caelifera. Recent data generation has uncovered another member of the family Tridactylidae that does not possess this rearrangement either.
Discussion
This study is this first attempt to reconstruct the phylogeny of an order of insects based on entire mtgenomes. We found that using mtgenomes in our phylogenetic analyses offered better resolution to our topologies than previous analyses and recovered a consensus phylogeny consistent with phylogenies based on morphological data (e.g. Eades 2000). Our consensus phylogeny supported the monophyly of Caelifera, while leaving the monophyly of Ensifera and Acrididae in question. Once we complete our data generation for Ensifera, we will be able to better test the monophyly of the suborder and its superfamily Tettigonioidea. Lastly, we found the tRNA rearrangement discovered by Flook et al. (1995) to be a useful phylogenetic marker. We found, however, that the rearrangement is a synapomorphy for the caeliferan group Acridomorpha (which includes all the caeliferan taxa in our taxon sampling except E. minuta) instead of for all of Caelifera.
References
- Eades DC. 2000. Evolutionary relationships of phallic structures of Acridomorpha (Orthoptera). J. Orthoptera Res. 9: 181-210.
- Fenn JD, Song H, Cameron SL, Whiting MF. 2008. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Molecular Phylogenetics and Evolution 49: 59-68.
- Flook PK, Rowell CHF, Gellissen G. 1995. The sequence, organization, and evolution of the Locusta migratoria mitochondrial genome. Journal of Molecular Evolution 41: 928-941.
- Kevan DKM. 1982. Orthoptera, Synopsis and Classification of Living Organisms. McGraw-Hill Book Company, Inc. p 352-383.
