Daniel Hammond and Dr. Brian Poole, Department of Microbiology and Molecular Biology
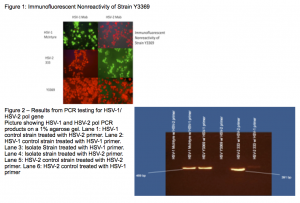
My project was focused on the identification and characterization of a clinical isolate of Herpes Simplex, the virus family responsible for cold sores and genital herpes. The virus was collected from a 48 year old female who was otherwise healthy. The patient had no previous history of genital herpes. The patient’s sexual partner was also HSV positive. The specimen was collected from a suspected infected genital tract and was submitted for analysis for Herpes simplex virus. Initial analysis using non-specific antibody tests verified that the virus was indeed a member of the herpes simplex family. Further analysis, shown in Figure 1, found the virus to be untypeable using the most common method for testing.
Through genetic testing I found the virus to be a variant of the Herpes Simplex 1 Virus. I used the Polymerase Chain Reaction or PCR to analyze the DNA of the virus at certain locations in its genome. I used primers whose accuracy had been shown previously in a peer-reviewed journal. The primers targeted a portion of the polymerase gene, the gene responsible for the virus’ own DNA replication, and are therefore highly conserved between different strains of the same virus. Figure 2 shows the results of the PCR that indicated the virus to be an HSV-1 variant.
After confirming the virus to be a variant of HSV-1 my research proposal indicated a genetic approach to test for different portions of different envelope proteins; the proteins that antibody tests target. I was able to successfully test multiple gene sequences for these proteins, however I was unable to find a sequence that was different up to now. After multiple tests Dr. Poole and I began to discuss other possible methods for discovering the reason why this particular strain was untypeable using antibody testing. As the virus was obtained from Dr. Johnson we consulted him, and together hypothesized that a very likely mutation was a frame shift deletion. That is to say, that the DNA polymerase did not copy a single DNA nucleotide and as a result the gene sequence was modified therefore causing vast changes in the protein product of a given DNA sequence. We will search for the protein by destroying an infected cell culture and through western blotting compare all the proteins present in a standard HSV-1 infection to the proteins present in our clinical isolate infected cell culture.
My proposal also included infection assays to determine possible differences in pathology between the two strains. Unfortunately I was unable to collect any verifiable data in this particular portion of my proposal, but it was still an important learning experience. I learned that live cell cultures are very delicate tools in scientific discovery. For the sake of my research I have been using vero cells, which grow very quickly, and if not cared for every two to three days, these cell cultures can quickly use all available food, and starve to death. In comparison the majority of the cell cultures in the Poole lab can go 5-6 days without receiving additional nutrients with no ill effects. In addition I learned that good science involves limiting the number of variables in each experiment. For example, when infecting a cell culture with a virus it is trivial to infect two equal volumes of cells, with equal volumes of virus. However determining if equal concentrations of cells were infected with equal concentrations of viruses is much more involved.
I consider myself fortunate to have made these errors. I had significant autonomy in carrying out these experiments. While this meant I made some rather elementary errors, I was able to learn from my own experience, and not simply follow the instructions of an established scientist. This suited my learning style better than any other learning experience in my undergraduate studies. In addition with the guidance of Dr. Poole I was able to find success in my project. The results of my research thus far were submitted to the Intermountain Branch of the American Society for Microbiology. At their meeting on April 10, 2010 I was selected for an oral presentation of my research. Many proposals were submitted, more than 50 were presented at the meeting, however only 8 were chosen for oral presentations. As an undergraduate I was given a rare opportunity. I would not have been capable of presenting in such a way had I not taken charge of this particular project through my ORCA grant proposal, and subsequent award.
Even though I have graduated I continue to work in the lab, despite the limited time I have as a result of full time employment. The pathology tests are nearing completion, and I continue to be involved in the research, although other members of the lab are carrying much of the time intensive work out. As a group we hope to be able to complete the infection assays, and protein analysis by the end of the year, and submit our findings to the Journal of Virology.