Taylor Gardner and Dr. Eric Wilson, Department of Microbiology and Molecular Biology
My purpose was to understand more deeply the process of mucosal immunological response in mice. Specifically, I hoped to understand the role of chemokine interactions in the recruitment of Immunoglobin A (IgA) in mouse colostrum/milk and mammary gland tissue during the gestation period. I focused on the time period just prior to the point at which milk production (and thus relative IgA levels) is upregulated in preparation for feeding young.
In order to provide myself an accurate reference point for analysis of the IgA response of the murine system I was required to be proficient in multiple lab techniques. Throughout my work with the murine subjects I also learned to approach scientific research and analysis in a systematic way. Keeping accurate records of all of my work was paramount, as there was a necessity for multiple test subjects, with each subject being sacrificed for multiple organs.
Before any tissues could be extracted I had to ensure a properly prepared test subject. As I was going to be studying the gestational murine mammary gland, I worked with Dr. Wilson in setting up matings of my required CCR9KO (A specifically changed mouse with a knockout of the CCR9 gene) mice. This did not prove successful originally, as none of the original matings produced any pregnant mothers. Upon further attempts, we were able to ensure that we had pregnant females.
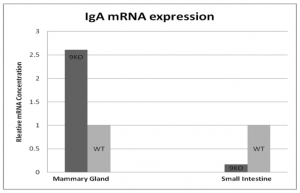
My hypothesis was originally that CCR9 was the determining gene in the transfer of innate immunity to the murine mammary gland in the pre-parturition period. As shown:
The information on low levels of IgA accumulation post day 21 are a result of previous research. My hypothesis was that virgin to Day 21(birthing) females accumulated these IgA levels via CCR9 interactions. Thus, with accurate controls and samples I would be able to understand better what interacts to produce an immunological response pre- parturition.
After I had sacrificed both CR57 (wildtype) and the CCR9KO females and collected their relevant tissues (mammary gland, small intestine) I used RNA extraction to produce samples of varying concentrations. I learned throughout this process that technique and accurate repetition are very, very necessary in a lab that is working with microliters and very small amounts of sample. I became, after a while, adept at the lab techniques for me to extract RNA from samples.
I went on to use the RNA samples from my WT and mutant strains to produce samples for qPCR analysis. An accurate understanding of qPCR techniques aided me in my learning of why and when I carried out my specific lab techniques. Using the RNA of different concentrations from my extractions, I set up qPCR reactions to begin finding results. This process was not one that went too smoothly. Indeed, many of my original attempts at qPCR were not effective. This was largely due to my errors in assembling the correct experimental set-up. In the middle of the process of collecting information from the qPCRs, Dr. Wilson’s lab purchased a PCR machine which I quickly learned to use (it differed in set-up from the machine that I previously used). This aided in the process.
After learning to accurately use the machinery, I was able to run successful qPCRs and produce real numbers and interpretations. When the original numbers had been analyzed I found that my hypothesis was not supported by my numbers. Indeed, it appeared that my CCR9KO mice had an increase in IgA concentrations when compared to the WT mice.
This phenomenon seemed to be tissue specific, as the small intestine samples tested seemed to show an opposite effect from that of the MG. This effect may be seen to reverse itself as more test subjects are included in data. The change may have been due to other factors, however. The possibility that some other migration aiding and inducing factor is contributing to the elevated levels of IgA mRNA transcripts is further supported by this data—yet the role of CCR9 in IgA ASC homing cannot yet be ruled out in the case of the pre-parturition Murine MG.
I was able to present these results at the American Society for Microbiology Intermountain Branch Conference. It was exciting for me to be able to share some of the work that I had put in on the subject of immunological response in the murine system.
Although there continues to be a necessity for further analysis via qPCR reactions and ELISA assays (for the protein samples that also can show IgA accumulation)—I feel that these preliminary results (and all the footwork I’ve done with collection and quantification of samples) are a significant steps toward understanding the importance of CCR9 in the homing of ASC in the gestational murine mammary gland. As a final note I found Dr. Wilson, my mentor, to be exceptionally patient and very helpful for me in my research. The experience was eye-opening and thoroughly successful in my education as a student of Molecular/Microbiology.