Bryan Merrill and Dr. Sandra Burnett, Department of Microbiology and Molecular Biology
Introduction
Honey bees pollinate one third of the crops in the U.S. diet. The spore-forming bacterium Paenibacillus larvae causes American Foulbrood (AFB), a highly contagious disease that is lethal in honey bee larvae. P. larvae is the most serious pathogen affecting honey bees. The spores can be found in any healthy beehive, are stable for 40 years or more, and cannot be killed by antibiotics. The disease manifests itself when worker bees feed P. larvae spores to larvae and the spores germinate. The bacteria then consume the larvae and turns back into a spore that the worker bees then transfer throughout the hive.
Antibiotics used to treat the disease are quickly becoming ineffective. Bacteriophages (viruses which infect and destroy bacteria) are a potential option to treat this disease. Although some P. larvae bacteriophages have been isolated, none of the genomes had been sequenced when this project began. Over the past year, five P. larvae bacteriophages have been isolated, sequenced, published, and analyzed. The information about these phage genomes marks an important first step in determining whether these phages can eventually be used as a treatment for AFB.
Materials and Methods
Local beekeepers donated honey and larva samples from both healthy and infected hives. Bacteria from these samples were grown on P. larvae-selective media (containing nalidixic and pipemidic acid). Bacterial colonies were confirmed as P. larvae by 16S rRNA testing.
Soil samples were collected near local beehives and then incubated in P. larvae bacterial cultures to propagate any phage in the soil sample. Phages were isolated by incubating P. larvae bacteria and phage samples together and then plating the mixture using the soft agar overlay method. Plaques produced by the phages were selected and purified three times. Solutions containing a high concentration of phages were used for transmission electron microscopy and 454 DNA sequencing. Genomic DNA sequences were then assembled, annotated, and published.
These bacteriophage genomes were analyzed in Phamerator, a powerful comparative genomics software program. This program analyzed 83 phage genomes, identified similar proteins between genomes, and grouped these similar proteins into “phams.”
Results and Discussion
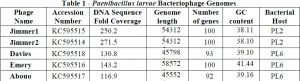
Fourteen P. larvae strains were isolated from donated samples and were numbered PL1 through PL14. BLAST analysis of 16S sequences suggested that all strains but one were P. larvae ssp. Pulvifaciens. Two of these strains, PL2 and PL6, were used to isolate phages. Of the four phage samples that were isolated, two infected PL2 and two infected PL6. No phages were shown to cross-infect the opposite strain. After DNA sequencing we found that one of the PL6 phage samples actually contained two distinct phage DNA sequences. These two phage genomes were assembled into single, complete sequences and named Emery and Abouo. Phage Abouo was used to assemble the three remaining phages. These phage genomes were annotated using DNA Master and submitted to GenBank for publication. Accession numbers for these phages are listed in Table 1.
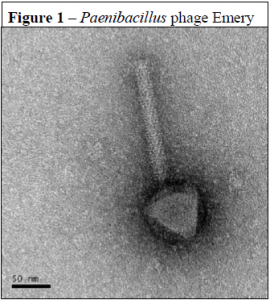
Transmission electron microscopy showed that all five of these phages have contractile tails and belong to the virus family Myoviridae (see Figure 1). Analysis of the physical structure and structural proteins of these five phages revealed similarity to Clostridium difficile phage PhiC2, Lactobacillus gasseri phage KC5a, and Streptococcus pneumoniae phage EJ-1 which are all small-genome myoviruses.
Phages Jimmer1, Jimmer2, Davies, and Abouo were more than 80% similar to each other while Emery was very different (less than 59% similarity to any of the other four phages). Jimmer1 and Jimmer2 are nearly identical (99.9% similar) and were isolated from the same soil sample. Similarities between these phages are shown in Figure 2 which is a portion of a Phamerator linear genome map. Based on Phamerator analysis of these and other bacteriophages, most of the genes in these five P. larvae phage genomes are novel. Only 23% of P. larvae phage genes were similar to other phage genomes that were analyzed. Further, only 42% of the P. larvae phage genes were found to contain a known conserved domain.
Conclusion
The five new P. larvae phages contain many novel genes and are not similar to any other phages that have been isolated and characterized. Now that these five phages have been characterized and published, there are three further goals: first, isolate and characterize more P. larvae phages; second, observe whether these phages can be transferred from adults to larvae in a beehive; and third, test whether P. larvae phages can successfully treat a hive infected with American Foulbrood.