JanaLynn Franke and Dr. Brad Geary, Plant and Wildlife Sciences
Introduction
With the accidental introduction of B. tectorum into the Western United States came a massive shift in the ecosystem, resulting in complete monocultures of B. tectorum. Seed and seedling pathogens present in the soil are aware of the macroscopic change and are shifting their populations as well in order to take advantage of the new abundance of food. One of these soil borne pathogens leads the way by manipulating the cheatgrass plant into a thick dense mat of litter, a perfect microclimate for seed pathogens that follow. This microorganism is most closely related to the fungal grass pathogen Roetstomia henningsiana Stromatinia gladioli, and Sclerotinea homeocarpa. This pathogen on B. tectorum infects at the crown of the plant and causes premature death of the plant which ultimately leads to the plants inability to set seeds. The purpose of this project was to more specifically identify this pathogen and to recreate the disease effects in a greenhouse experiment.
Materials and Methods
DNA Sequencing
Previously collected fungal isolates from the Great Basin were re-sequenced using freshly extracted DNA (ZR Fungal/Bacterial DNA miniprep kit). A PCR (Polymerase chain Reaction) was done using the primers ITS1 through ITS7. Once amplification was achieved samples were purified and sequences were run through Big Dye and results were obtained. The sequence results were then aligned and blast using the computer program Geneious.
Greenhouse experiment-inoculum production
The fungal isolate previously collected was grown in ½ strength PDB (Potato Dextrose Broth) for 1 week. A chaff grain mixture was used at a ratio of 5:1. The chaff grain mixture was then soaked in water for 24 hours to allow leaching of phenolic compounds. Mixture was strained and placed in autoclavable glass jars and cotton placed over the mouth of the jar. The Jar and mixture were then autoclaved twice for 40 min. each. After the mixture was cooled it was inoculated with the PDB culture and a freshly autoclaved cotton piece placed back over the mouth of the jar and secured. The jar with inoculated mixture was finally incubated at 25ºC until the fungus had adequately colonized the mixture. Mixture was dried, ground up and 1 gram per plant weighed out.
Greenhouse experiment-set-up
Non-dormant B. tectorum were cleaned and planted into a low nutrient soil. After planting plants were randomly divided into thirds. The first third were used as a control. Then the second 1/3 of the seeds were inoculated with the fungus for the first treatment. All plants were watered and placed in a cold room to vernalize. After 3 weeks of vernalization the seeds sprouted and the last 1/3 of the plants were inoculated for the second treatment. All the seeds were then moved to a greenhouse and kept at a temperature of 25-35ºC for the remainder of the experiment. Shallow watering happened 1-2 times a week to ensure a lower water potential. Finally plants were harvested and the plant and seed biomass weighed and seeds counted per plant. Crown samples were taken from infected plants and cultured onto PDA.
Results and Discussion
Sequencing results
Blast results were inconclusive and did not shed any more light on the genetic identification of this isolate. Based on previous sequencing data genetic relation goes as follows Rutstroemia henningsiana, Stromatinia gladioli, and Sclerotinia homeocarpa.
Greenhouse Experiment
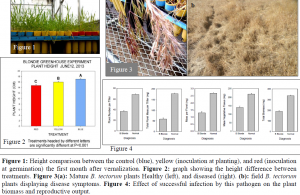
Within the first month of growth, a definite height difference became apparent as seen in Figure 1. After measurement of each plant results showed that height was statistically different between treatments. Due to an accidental increase in watering for a few weeks, heights differences between treatments decreased towards the end of the experiment. However, symptoms of the pathogen did not completely disappear. Once the watering was corrected and the B. tectorum plants matured the symptoms reappeared, manifest in bleach blond florets as seen in Figure 3a. This bleach blond color is indicative of this pathogen when seen out in the field (Figure 3b.) This pathogen’s symptoms were also apparent in the biomass weights. The most common symptom used to identify this disease is the lack of seeds filled. Our results were conclusive, and supported symptoms seen in the field. As seen in Figure 4, all aspects of the plant were affected from quantity of seeds per plant to the biomass of the plant. According to all our results this pathogen is able to infect the plant while in the seedling stage and slowly cutoff its food supply at the crown. This results in a plant that prematurely tries to set seed but ultimately fails.