Loyd Christensen and Dr. Laura C. Bridgewater, Micro and Molecular Biology
Introduction
Intervertebral Disc Degeneration (IDD) is a disease estimated to affect between 60 and 90 percent of the total population at some point in life. Evidence of spinal disc degeneration has been shown in 30 percent of individuals under 40 years old, with earliest symptoms visible at 21 years of age; its prevalence only increases with age. Symptoms of spinal disc degeneration account for more than 13 million doctor visits each year and result in costs of more than $200 billion in lost productivity and health-care related expenses in the US alone. Unfortunately, despite these staggering costs, no widely effective treatment has been developed.
The study of IDD is hindered by generally recognized limitations of currently available animal models. First, most models rely on injury based methods such as annular stab or injection of matrix proteases to trigger degeneration, which may not accurately model spontaneous IDD in humans. Second, currently available model animals have horizontal rather than vertical spine configuration.
Fortunately, alpaca vertebrae have been shown to be effective models for the human spine based on similarities in anatomical shape, flexibility, and mechanical load distribution. Knowing this, we studied genetic aspects of alpaca spinal discs to determine the relative usefulness of alpacas as a biological model for human IDD.
Methodology
To test the biological viability of an alpaca animal model, we used both Live/Dead staining and quantitative Real Time-PCR (qRT-PCR) amplification of alpaca cDNA.
To establish post-excision tissue viability, we excised discs from the vertebral column, cleaned and cultured them in D-MEM in a custom-built bio-reactor for one week. We then stained the discs with propidium iodide (which stains dying cells red) and with CyberGreen (which stains live cells green).
To determine the level of homology between alpacas and humans, we used human-specific qRT-PCR primers to measure the expression of Aggrecan, Col2a1, Col1a1, SOX9, MMP13 and ADAMTS5 genes in alpaca intervertebral discs.
Results
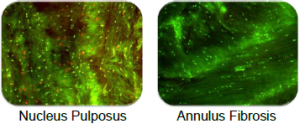
Live Dead Staining
As shown above, nucleus pulposus cells showed some cell death (red stain) during culture. This is arguably due to the difficulty nutrients face in crossing the dense outer fibrous layer. Annulus Fibrosis cells were, on the other hand, mostly alive (green stain).
![]()
We also used human-specific qRT-PCR primers to measure the expression of Aggrecan, Col2a1, Col1a1, Sox9, MMP13, and ADAMTS5 genes in alpaca IVDs. There is enough homology between the two species to successfully amplify each of these genes involved in cartilage formation and to merit further development of alpaca specific qRT-PCR primers.
Discussion
The above results point to the alpaca as a potentially valuable model organism. The ability to maintain viable tissue in vitro is foundational for future studies to test the impact of mechanical loads on spinal disc composition. Further, the ability to amplify alpaca cDNA with human qRT-PCR primers points to a significant level of homology between the two organisms and is indicative of future success of further quantitative genetic studies
Conclusion
The alpaca is a valuable model organism for the study of intervertebral disc degeneration. This project completed essential preliminary work toward the ultimate goal of reversing IVD degeneration and improving the quality of life for thousands of individuals.