Karl Andersen and Dr. David Kooyman, Physiology and Developmental Biology
Osteoarthritis increasingly debilitates and causes discomfiture to millions of people worldwide. Most people over 65 years of age suffer from symptoms of osteoarthritis. There has, up to this point, been effectively no clear understanding of the processes of the body that contribute genetically or mechanically to this severe condition. The result of this lack of knowledge is a general absence of prevention and treatment measures. As of today, treatments consist only of joint replacement and pain reduction measures.
Dr. Kooyman and his lab have been studying the molecular progression of osteoarthritis for four years. Based on his work and that of others, this project has attempted to determine whether TgfB1, acting as a cytokine/growth factor, upregulates HtrA1 expression and contributes consequently to the induction of osteoarthritis. This work-‐in-‐progress has given results on expression in wild-‐type mice. Our experimental model consists of a comparison of expression between normal, wild-‐type mice and an SEDC (Spondyloepiphyseal Dysplasia Congenita) model that is genetically predisposed to osteoarthritis. We have completed the proposed research in part by obtaining results for the wild-‐type mice, and we will continue experiments to elucidate expression in the SEDC model and make the resulting comparison.
We bred mice colonies in accordance with BYU IACUC (Institutional Animal Care and Use Committee) policies and guidelines. At varying ages, mice tissues were harvested with a method approved by IACUC protocol 10-‐1102. Hip cartilage was harvested from mice under isoflurane anesthesia, and after a humane sacrifice procedure, synovial fluid was also retrieved from mouse knee joints. Hip cartilage and synovial fluid provided samples on which we ran our experiments. To determine quantitative protein presence of TgfB-‐1 in these samples, we used an ELISA (Enzyme-‐Linked Immunosorbent Assay) technique. To age-‐matched samples, we also performed gene expression analysis using qPCR (quantitative Polymerase Chain Reaction). This data allowed us to compare mRNA levels with realized protein levels in the hip cartilage of mice. Samples were stored in saline solution at –20° Celsius for samples used in the ELISA and –80° Celsius for samples used in qPCR. In order to prevent mRNA degradation, we used a snap-‐freeze technique by placing hip cartilage samples on liquid nitrogen almost immediately after harvesting.
For the ELISA, we used a kit ordered from Invitrogen. After thawing and homogenizing the samples, we followed the protocol included in the kit. Finally, to obtain quantitative levels of the TgfB-‐1 protein, absorbance readings were taken of purified protein samples using a spectrophotometer. For qPCR analysis, mRNA was stabilized while thawing using Trizol solution. Tissues were then homogenized and RNA was purified and isolated using a Trizol RNA isolation kit.. Using a Nanodrop machine, we then verified mRNA purity. Each sample was then converted to cDNA using RT-‐PCR. The cDNA kit and accompanying protocol were 0 100 200 300 400 500 600 700 800 10 weeks 12 weeks 24 weeks Protein Presence pg/ml Synovial bluid Hip cartilage also obtained from Invitrogen. cDNA samples were then subjected to real-‐time PCR, and RNA levels were quantitatively measured with a fluorescent dye and a melting curve.
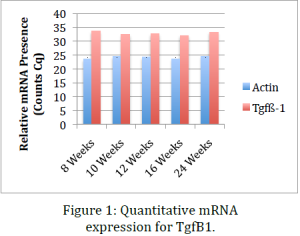
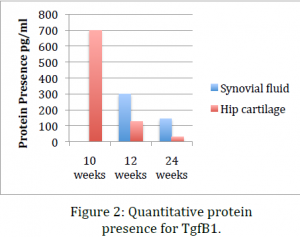
Data obtained from the before mentioned procedures indicate that, while RNA levels for TgfB1 remain relatively constant throughout the life of the mouse, protein presence significantly drops with age and the onset of osteoarthritis. This indicates that one explanation might be that the inflammation response or pathway does not inhibit transcription of TgfB1 genes but rather inhibits translation of mRNA or stops post-‐ translational modifications that render the protein products useless. This data coincides with data previously obtained in our lab using immunohistochemical stains of sections of knee tissue. Dr. Kooyman’s lab has already shown that in mid-‐stage and late-‐stage osteoarthritis TgfB1 is significantly down regulated. The ELISA we performed in this project confirmed this conclusion using an alternative method. This provides a second evidence that cellular growth factor TgfB1 is indeed downregulated in osteoarthritis.
The procedures performed in these experiments have shown the gene and protein expression of TgfB1. When we compare this data with HtrA1 expression, in future work, we will be able to ascertain whether a relationship exists between HtrA1 and TgfB1 in which the latter upregulates the former and destines cells in articular cartilage to apoptosis (cell death). This clarified understanding of the inflammation pathways in arthritis will move the understanding of modern science one step closer to the prevention and treatment of osteoarthritis on a molecular level.
Future work will include another ELISA on wild-‐type samples as well as qPCR and ELISA procedures performed on samples from SEDC mice. This research has been submitted for an oral and/or poster presentation at the Experimental Biology 2014 Conference in San Diego scheduled for April 26-‐30. This project will also provide the bulk of my advanced senior research project that will result in a scientific article submission to Osteoarthritis and Cartilage at the end of April 2014.