Steven Nevers, David Brandt, and Matthew Biggs
1 Abstract
Despite previous work by our group and other researchers, the existing methods to predict ADAMTS13 deficiency are inadequate. Through the use of machine learning techniques applied to mined patient data we have improved the accuracy and efficiency of our existing ADAMTS13 deficiency prediction algorithm. In conjunction with ARUP Laboratories, who pioneered the original work, we have continued to improve the algorithm accuracy. Significant correlation for severe ADAMTS13 deficiency was seen for four of the observed variables: indirect bilirubin, reticulocyte percentage, creatinine, and platelet count; a fifth variable, D-dimer, just missed significance but performed well.
They are listed in order of decreasing correlation. We utilized the software program Weka1 to train and test our algorithm on the datasets. The results of our machine learning showed that the algorithm correctly classified patients 90.2% of the time (185/205) and incorrectly classified 9.7% of the time (20/205).
2 Introduction
Idiopathic thrombotic thrombocytopenic purpura (TTP) is a disorder of the blood-coagulation system that causes microscopic clots to form in the small blood vessels throughout the body. It is characterized by an extreme deficiency of the enzyme ADAMTS13. In previous work we attempted to identify factors that can be used as an indicator to predict ADAMTS13 deficiency. The test for ADAMTS13 is expensive and must be outsourced to a reference laboratory. Being able to predict deficiency with only a handful of standardized lab tests would decrease medical costs. Over the course of the last 9 months we have: (1) validated the previous algorithm against a new cohort of data, (2) improved a decision tree algorithm to predict the probability of a patient being ADAMTS13 deficient, and (3) created a neural network as an additional learning technique in an attempt to improve the algorithm, and (4) utilized Weka to test and train to develop an algorithm.
3 Related Work
We could not find specific published work that involved an algorithm to predict deficiency. Most of the articles are focused more on chart-reviews that give ranges of lab values for patients with ADAMTS13 deficiency. The French TMA Reference Center recently published the results of their chart-review and their computed values.2 The only work that is published in regard to a prediction algorithm was the one published by our mentor entitled: “The utility of patient characteristics in predicting severe ADAMTS13 deficiency and response to plasma exchange”3.
4 Materials and Methods
For our previous project we improved an existing decision tree. We started with a large collection of patient data.
Previous work on this project has shown 5 factors that are the most significant. We ran an initial logistical regression permuting the data to attempt to validate these 5 factors on the new data. They all were significant again. Now that we have confirmed the significant ones we ran a full logistical regression to determine the strength of each predictive factor. From this output we generated a decision tree.
As an initial step for improvement we developed a neural network. A neural network is a computational model that is inspired by the structure and functional aspects of biological neural networks. A set of data can be encoded into the network. Specifically, in supervised learning we give the program a set of examples and it attempts to find a function in the classes of data that matches the examples. It involves an input layer, a hidden layer, and an output layer.
Between each is a series of weights that determine the propagation of a signal. A signal propagates from the node that matches the data pattern.
The second attempt for improvement was using the machine learning software Weka. It is a collection of visualization tools and algorithms for data analysis and predictive modeling.
For some of the patients we were missing data values. To correct for this we used the nearest neighbor algorithm. This allows us to guesstimate a missing values based on closest training examples.
We attempted several different built in tests. The best performing was the 100-fold cross-validation. Weka took a sample of the dataset and use it to train and then test it on the remaining unused part of the dataset. Then it will do this 100 times over and over taking different samples.
5 Results
We improved our predictor algorithm but not to a level of significance that would be necessary to be used in a clinical setting. Among the different improvements attempts the results generated by Weka were by far the best. All of the training and testing was done on the 205 instances. One of the problems with machine learning is that with small data sets it is easy to train your algorithm to your data and it will perform well. But when it is used on new data it hasn’t seen before it performs poorly. Weka’s method of dealing with this is training and testing on different subsets of the data over and over. This helps remove this pseudo training. The most successful algorithm based on the data sets was very successful. It correctly classified 90.2% of the instances (185 instances out of 205 total) and incorrectly classified 9.7% of the instances (20 instances out of 205 total). This was significantly better than our previous algorithm.
6 Conclusions
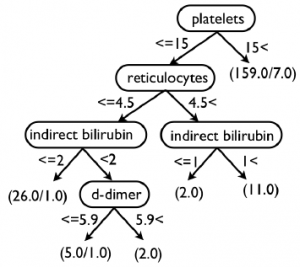 Fundamentally, the level of predictor we are getting and the success of training is good based on the small amount of data we have. In order to develop a more robust and sensitive predictor we need to have more data sets to train the algorithm on. Out of 205 patients we only had about 30 individuals with TTP and so that small percentage makes it difficult to predict. Future work may continue with other researchers that are also working on similar projects to share data and help get better results.
Fundamentally, the level of predictor we are getting and the success of training is good based on the small amount of data we have. In order to develop a more robust and sensitive predictor we need to have more data sets to train the algorithm on. Out of 205 patients we only had about 30 individuals with TTP and so that small percentage makes it difficult to predict. Future work may continue with other researchers that are also working on similar projects to share data and help get better results.
Works Cited
- Mark Hall, Eibe Frank, Geoffrey Holmes, Bernhard Pfahringer, Peter Reutemann, Ian H. Witten (2009); The WEKA Data Mining Software: An Update; SIGKDD Explorations, Volume 11, Issue 1.
- Coppo P, Schwarzinger M, Buffet M, Wynckel A, Clabault K, et al. (2010) Predictive Features of Severe Acquired ADAMTS13 Deficiency in Idiopathic Thrombotic Microangiopathies: The French TMA Reference Center Experience. PLoS ONE 5(4): e10208. doi:10.1371/ journal.pone.0010208
- Bentley, M. J., Lehman, C. M., Blaylock, R. C., Wilson, A. R. and Rodgers, G. M. (2010), The utility of patient characteristics in predicting severe ADAMTS13 deficiency and response to plasma exchange. Transfusion, 50: 1654–1664. doi: 10.1111/j.1537-2995.2010.02653.x Final
