Scott Smith and Dr. Jeff Maughan, Plant and Wildlife Sciences
The grain amaranths (Amaranthus sp.) are important pseudo-cereals native to the New World. During the last decade they have garnered increased international attention for their nutritional quality, tolerance to abiotic stress and importance as a symbol of indigenous cultures. Notwithstanding their renewed potential as an emerging alternative crop species, few molecular tools needed for accelerated breeding programs, have been developed. Our research describes the development and characterization of the first 411 single nucleotide polymorphism (SNP) marker assays for the species as well as the first complete genetic linkage map of the species.
SNPs were developed by extracting and comparing the DNA of two individuals, then identifying single nucleotide polymorphisms between them through the use of software developed at BYU. A subset of 500 of the identified SNPs were then developed into KASPar™ genotyping assays which when screened, should identify the allele at each marker based on clustering using the Fluidigm access array system (Fig. 1). Once developed the markers were screened on an F2 population of 134 individuals from an interspecific A. hypochondriacus X A. Caudatus cross. Of the 500 assays developed 411 produced clearly separated genotypic clusters that could easily be scored. The genotypic data of these markers was used to create a genetic linkage map using Joinmap (Fig. 2). The map consists of 16 groups of markers that tend to be inherited together, presumably representing the 16 chromosomes of amaranth. The distribution of markers range from 9 to 47 markers per linkage group spanning a total of 1288 centiMorgans (cM) with an average distance between markers of 3.1 cM. The SNP markers and linkage map we developed are the initial first steps towards the genetic dissection of agronomically important characteristics in amaranth.
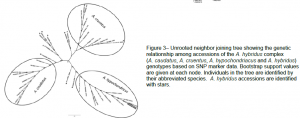
In addition to screening these markers on an F2 population we also screened them on a diversity panel of 41 individuals composed of the 3 domesticated grain amaranths and their putative wild ancestor A. Hybridus. The genotypic data from the diversity panel was analyzed phylogenetically in order to see which individuals are most closely related to each other. The results showed that the 3 grain amaranths A. caudatus, A. cruentus and A. hypochondriacus are monophyletic, while A. hybridus was polyphyletic with A. hybridus accessions in each of the three grain amaranth monophyletic clades (Fig. 3). These results support the designation of A. hybridus as the progenitor species of all three grain amaranth species. Our phylogenetic analysis also clearly places one accession within the A. hypochondriacus clade which is designated as A. Caudatus within the GRIN system, suggesting that this accession was originally misclassified.
The results of this study were presented at the 2011 Plant and Animal Genome Conference in San Diego in the form of a poster. In addition to this presentation The full results were also published in the peer reviewed journal The Plant Genome (Maughan, 2011).
Figures
References
- Maughan, P., Smith, S., Fairbanks, D., & Jellen, E. (2011). Development, characterization, and linkage mapping of single nucleotide polymorphisms in the grain amaranths (amaranthus sp.). Plant Gen., 4(1), 92-101.