Robert Nielsen and Dr. Craig Colemon, Plant & Wildlife Sciences
FLOWERING LOCUST like (FTL) genes are crucial regulators of flowering in angiosperms. Recent discoveries have shown that flowering in young seedlings of C. rubrum, a model plant for the study of photoperiodic flower induction, and a close relative of C. quinoa, is highly inducible by a 12-hour dark period. I sought to measure how changes in expression of genes associated with flowering time are correlated with the transition from a vegetative state to an active flowering in the short-day plant C. quinoa. Understanding C. quinoa may one day enhance our knowledge about plants that flower in low-light environments and lead to improved growing methods.
A great deal of speculation into this field has grown its popularity, and I had the opportunity to collaborate with a few great names in the field of photosynthesis research, namely Helena Storchova, with whom I was privileged to work with for three months in Czech Republic, and Alejandro Bonifacio, from the McKnight Foundation’s Collaborative Crop research program. Our work may one day open a pathway to far more effective methods of plant breeding and production in countries such as Peru, Bolivia, and Ecuador, where the increasing popularity of Quinoa as an export grain has left much of the population undernourished and under fed.
Signaling pathways receptive to environmental cues are responsible for the accurate regulation of flower induction (reviewed by Lee et al. 2006). The genes I studied had all been identified in A. thaliana, and their homologs exist in C. quinoa.
Although the same genes are present, we found that they are expressed in different patterns, which may account for the difference in short day and long day induction of flowering. We used studies done on CrFTLJ and CrCO orthologs in C. rubrum to show that indeed the gene expression pattern of these two genes is different than what is seen in A. thaliana under floral inducing conditions.
To do this we examined the differences that existed in the expression pattern of the CqFTLJ and CqCO genes in C. quinoa. We initially ran into some difficulties while isolating the gene, which we found to be elusive because of its relatively low GC content. Formulating primers to capture the sequence was a vital first step in the capture and analysis of the sequence of CqFTLJ and CqC. Our findings required a few repetitions to get the needed results.
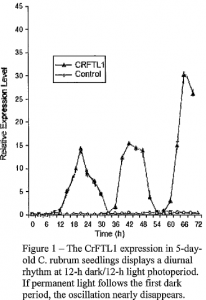
I subsequently used qRT-PCR to measure the expression of FTLJ and CO homologs in cotyledons and the first leaves of plants with reduced inducibility in C. quina a (7- 14 days old) (Seidl ova and Krekule 1973; Ullmann et al. 1985). Our fmdings were not far different from that present in C. rubrum, which we determined to be logical based on their sequence homology. Flowering could be induced by a single 12-hour dark period in five day-old seedlings of C. quinoa.
I also tested whether or not FTL genes can be expressed at high levels in cotyledons, but not in young first leaves or mature cotyledons (> 7 days old). Data showed that flowering can be induced by a single 12-hour dark period in five day-old seedlings of C. rubrum, however, this permissive developmental period is followed by a period ofreduced photoperiodic sensitivity (Seidlova and Krekule 1973; Ullmann et al. 1985). Via qRT-PCR I measured the expression of FTLJ and CO homo logs in cotyledons and the first leaves of plants with reduced inducibility in C. quina a (7 -14 days old). My findings, demonstrated in figure 1, reflect that the hypothesis was correct; flowering can be induced by single 12-hour dark period followed by a period of reduced photoperiodic sensitivity. The diurnal activity of the plant reflected the distinct changes in RNA expression of FTL genes.
I believe that these genes do in fact play a major role in growth regulation in C.quinoa and serve as one of the leading measures of its viability. Future efforts will focus on how these control mechanisms can be regulated and managed through experiments in bisulfite sequencing. All in all, I am deeply grateful for the funding that contributed to the success of this project.