Cecily P. Vaughn and Dr. Randy Bennett, Zoology
Development in animals is controlled by a set of genes called homeotic genes whose protein products regulate the expression of other genes. Although many homeotic genes have been locates in numerous organisms, there is still much that remains to be learned about the manner in which these genes control development. The aim of my ORCA project was to determine the expression patterns of a few key developmental genes in the red flour beetle (Tribolium castaneum).
I used a method called in situ hybridization to look at the expression patterns of developmental genes in the developing embryos of Tribolium. In situ hybridization leaves the organism intact throughout the probing and staining steps so that the exact location of gene expression can be observed.
The first step in my project was to collect beetle embryos at various stages of development. The next step entailed fixation of the embryos, which was done to halt their development and prepare them for staining. Part of my project involved testing the different methods of fixation to determine which produced the best results. Although it is more time consuming, the chemical method was chosen over sonication because it resulted in fewer damaged embryos and embryos which were more susceptible to staining.
The next step involved attaching probes to the products of the genes of interest and then staining the probes so that the gene expression was visible. For most of the genes antibody probes were available. For the main gene of interest, Ultrathorax (Utx), the antibody has not yet been made, so I had intended to use an RNA probe. When I wrote my research proposal I had anticipated that I would be able to obtain the Utx RNA probe from another lab. This however was not possible and so I had to prepare the RNA probe myself. This presented the major delay of my project because the probe had to be prepared before I could do any stainings for Utx expression. I prepared the RNA probe using a DNA clone of the Utx gene. I linearized a plasmid containing the DNA clone and then incubated it with a kit that contained ribonucleotides and RNA polymerase. The polymerase synthesized a piece of RNA using the DNA as a template. The RNA that I then isolated was complementary to the mRNA product of the Utx gene.
Each staining involved several days. The embryos were first incubated in a tube with the probe, either an antibody or the Utx RNA probe. They were then washed numerous times. In most cases a secondary antibody was used, followed by an enzymatic reaction that produced a color change, marking the places where the protein was present. For the Utx RNA probe a stain specific to the nucleotides in the RNA probe was used to stain for the gene expression.
After the staining I dissected the embryos. Each embryo is about the size of the tip of a ballpoint pen, so all of the dissecting had to be done under a microscope. An intact embryo is surrounded by yolk and enclosed in an outer membrane. I had to remove the membrane and scrap the yolk away from the embryo. This process resulted in many torn and therefore unusable embryos, so dissecting was a skill that I spent many hours practicing. The embryos that are successfully dissected were then mounted on slides and color pictures were taken through the microscope.
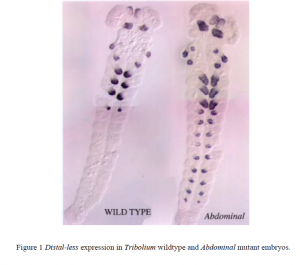
In my proposal I had proposed to do stainings to study the interaction of the Utx gene and the hunchback gene. I had also proposed to study the interactions of Utx and Abdominal with Distal-less (Dll), a gene that is responsible for limb development. In the studies that have continued in the lab this summer, it has been observed that Utx does not turn off the Dll gene, as it does in Drosophila, but that Abdominal does turn off the gene, just as it does in Drosophila. The fact that Abdominal turns off the Dll gene was observed by staining wildtype embryos and Abdominal mutant embryos. Compared to wildtype embryos, Distal-less expression extents further posteriorly in the Abdominal mutants.
The techniques that were necessary to develop and perfect took much longer than I had anticipated when I wrote my proposal. Although there were many setbacks that I encountered, the information that I obtained by working through the problems will be useful as the lab continues the work on developmental genes in Tribolium.