Joseph A. Garcia and Dr. Terry S. Elton, Chemistry and Biochemistry
The study of human genetics has recently exploded into numerous fields and, as a result, our knowledge of genetics and gene related illnesses is growing exponentially. Human DNA undergoes three main levels of control. The first is replication where the genome is reproduced in an exact copy. The second level is transcription, where an RNA polymerase transcribes DNA into mRNA. Finally translation where mRNA, the code for making a protein is recognized by ribosomes. Ribosomes recognize the 5′ region of the mRNA and scan along until they find the start codon to initiate the translation of the mRNA into a protein. This is called the cap dependant mechanism. My research will focus on the translational control of the human angiotensin II type 1 receptor (hAT1R) and how the mRNA is specifically recognized by ribosomes. The hAT1R receptor mRNA translational regulation has serious implications in the role of hypertension and can also be linked to cell growth and indirectly to tumor neovascularization.
The hormone Angiotensin II (AngII) plays a crucial role in physiological and pathophysiological cell growth and angiogenesis. Ang II binds to and induces cell growth for several types of cells; these include vascular smooth muscle cells (VSMC), renal mesengial cells, cardiomyocytes and cardiac fibroblasts.
Ang II binds to a G-protein-coupled receptor on the target cell surface. Two types of the Ang II receptors have been identified, ATIR and AT2R. When Ang II binds to AT1R it activates a cascade of events. First it stimulates phospholipase C which cleaves specific phospholipid residues leading to the production of a triphosphate and a glycerol molecule. The presence of these two molecules are involved in intracellular Ca 2+ mobilization and protein kinase C activation. AT1R activation also stimulates another cascade of events known as the ERK 1/2 cascade. This cascade has recently been linked to Shc-Grb2-SOS complex formation and RAS activation, which leads to the activations of ERK 1/2. This pathway can be linked to cell growth regulation and may indirectly be linked to cancer development in certain tissues. The AT2R pathways are not well defined but have been linked to inhibition of proliferation and may antagonize growth promoted by AT1R.
It has been demonstrated by Dr. Elton’s (my mentor) laboratory as well as others that the human AT1R (hAT1R) gene has at least four exons and spans 60 kilobases (kb) or greater. Exons 1,2 and 3 make up the 5′ untranslated region (5`UTR) mRNA sequence. Exon four contains the entire open reading frame for the hAT1R.
Very little is known about the possible translational control of the AT1R mRNA. However, the 5′UTR contains important structural clues, exons 1,2 and 3 are very GC rich (78.4%), which means that it can form very stable secondary structures. Additionally the 5′ UTR contains an AUG triplet upstream from the actual translational initiation start codon. These features would impose a significant barrier to the scanning method mentioned before, suggesting that hAT1R mRNAs may be translated by a “cap-independent” translational mechanism.
Like the hAT1R mRNAs, other regulatory genes have been observed to have these unique characteristics in their 5′UTR these include, fibroblast growth factor 2 (FGF-2). Insulin-like growth factor II (IGF-II), platelet derived growth factor (PDGF), vascular endothelial growth factor (VEGF) and the c-myc proto-oncogene. All of the 5′UTRs of the mRNAs of these genes possess internal ribosomal entry sites (IRES) and can be translated by a cap-independent mechanism.
Translational Regulation Through an Internal Ribosome Entry Site Present in the Human Angiotensin II Type 1 Receptor mRNA.
Many mRNAs encoding growth factors, cytokines and oncoproteins are read via their IRES element. These IRES elements may provide translational regulation mechanisms in response to changes in physiological conditions. Because these IRES elements are found in growth factors, this may suggest that the processes leading to cell transformation and tumor neovascularization could involve regulation of IRES dependent activation of the expression of these factors.
Dr. Elton’s (my mentor) lab demonstrated that the hAT1R receptor might contain an IRES element, which has implications that the hAT1R is also translationally regulated like many of the other growth factors. It is my goal to investigate the functional importance of this sequence and to determine if the hAT1R is translationally regulated, therefore I will characterize the IRES element by,
AIM 1. Identifying sequences within the 5′-UTR of the hAT1R mRNA required for IRES activity.
AIM 2. Identifying trans-acting factors that recognize the IRES sequence.
Procedure
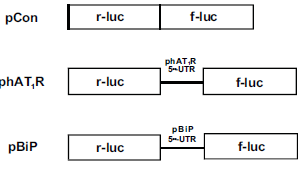
AIM 1. To identify the exact sequences in the hAT1R 5′UTR required for IRES activity, hAT1R 5′ UTR bi -cistronic plasmids will be constructed. Portions of the hAT1R (exons 1-3) will be PCR amplified and sub-cloned between two cistrons. The new constructs will be sequenced to verify their orientation and then transfected into Human Adrenocortical Carcinoma cells (H295-R). The constructs will contain a reporter gene firefly-luciferase (f-luc) followed by a region of the 5′UTR of different lengths that is then followed by renilla luciferase (r-luc). The reporter genes will be expressed proportionally to how well the 5′UTR region of varying lengths induces expression. Dual luciferase assays shall be performed and the ratio of f-luc/r-luc will be compared to the luciferase ratios obtained from H295-R cells transfected with phAT1R. The phAT1R control construct has the entire hAT1R 5′UTR subcloned between the two cistrons, whereas the others have varying lengths of the 5′UTR region. All data obtained will be compared to a control bi-cistronic harboring the 5•-UTR of the known IRES from the binding immunoglobulin protein (BiP) mRNA (Fig. 1). The integrity of the bi-cistronic mRNA will be verified by Northern blot analysis using 32P labeled cDNA probes specific for r-luc and f-luc.
As different regions of the hAT1R 5′UTR are deleted and compared with the entire 5′ UTR sequence it is expected that the exact location of the IRES will be located. It is expected that the f-luc and the r-luc ratio will decrease significantly if the region of the IRES has been deleted in any form. Upon finding the location of the IRES in the 5′UTR, its sequence will be analyzed to find the thermodynamically favored helical stems, which the ribosomes will recognize, (34).
As was suspected, a 100 base pair deletion, the H295R cell transformation resulted in an eight fold decrease in luciferase expression compared to that of Bip which has known IRES activity. (See chart below). When the 1b in 4 construct was left without deletions the H295R cells showed a 14-fold increase over Bip. We believe that the IRES site is within this 100 base pair region due to the GC secondary loop structure that it is capable of forming. Our results were also confirmed by cycle sequencing to ensure constructs position in construct and base pair accuracy as compared to in vivo expression. It remains to be determined whether this site is an actual promoter site or an IRES site.
Figure 2 Done with the help of Mickey Martin 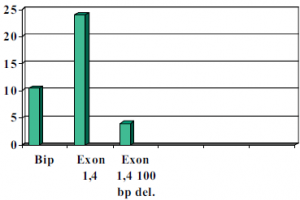
Much of our time was spent making correct and accurate constructs. Tests will be done in the near future to determine whether the exons 1,4; 1,2,4; 1,2,3,4; 1,3,4 have IRES activity by the same method. Most of these constructs have already been made and the results will be published in a separate journal.
AIM 2 Results: Time did not permit us to reach this part of the experiment. This experiment will be done upon completion of all constructs and the results will be published in another journal.
Special Thanks To:
Terry Elton, Mickey Martin, Xin Zhao and Xylophone Victor.