Susanna M. Geary and Dr. Roger Kaspar, Chemistry and Biochemistry
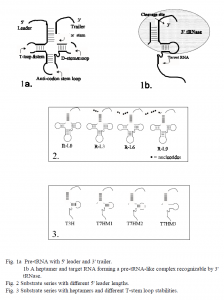
Viruses insert DNA or RNA into host cells and use the cell’s resources to make proteins designed to propagate the virus. Viral proteins can cause disease, as can defective proteins from faulty or damaged genes. The enzyme, tRNA 3′ processing endoribonuclease (also called 3′ tRNase) can specifically cleave RNA that is made to resemble pre-tRNA (1). RNA heptamers can be engineered to form such pre-tRNA-resembling complexes by binding to a target RNA just downstream of a stable hairpin loop (fig. 1). Hence, pathogenic RNA could be targeted and destroyed, thereby decreasing the production of damaging proteins.
In attempt to define substrate structures that would maximize 3′ tRNase cleavage efficiency, I analyzed two sets of potential substrates: the L0, L3, L6, L9 series consisted of arginine tRNAs, identical except for 5′ leader length (fig. 2); the T3H, T7HM1-3 series consisted of target RNA/heptamer complexes, identical except for T-stem loop stability (fig. 3). Secondary structures were assessed by labeling each series with radioactive (32P) nucleotides and then digesting them with RNases A, T1 and V1. Resulting fragments were separated by size in a polyacrylamide gel and detected by autoradiography. Relative kinetic parameters of 3′ tRNase cleavage efficiency, Vmax/Km*Kd, were obtained for each series by kinetic assays.
Structure probing results of the T3H, T7HM1-3 series were inconclusive— most likely because of the equilibrium of attaching and detaching heptamers. However, the free energy ( G) of T-stem loop formation for each complex was calculated and were approximately proportional to the kinetic parameters, indicating a direct correlation between T-stem loop stability and enzyme cleavage efficiency. For example, T3H had a G of -5.2 kcal/mol, and a relative kinetic value of 1.0; T7HM1 had a G that was approximately half the value of T3H’s (-2.3 kcal/mol) and a relative kinetic value also approximately half that of T3H’s (0.6).
The structure probing assays on the L0, L3, L6, L9 series indicated that despite the differences in 5′ leader length, each substrate had identical secondary structures; any difference in cleavage efficiency would be a direct result of 5′ leader length. Apart from two bands on the autoradiograph, which were the result of the 5′ leader cleavage, there was no distinction between the L0 and L9 digestion patterns. Furthermore, 5′ leaders of samples L9 and L6 were digested by RNase T1, which indicated that these leaders were single stranded and therefore could not have been involved in double-stranded secondary structures with the rest of the molecule. Kinetic assays indicated that cleavage efficiency increased as the 5′ leader length increased. This trend held for the first three samples (L0, L3 and L6), but was broken with L9, since it was not cleaved at all (2).
References
- Nashimoto, M., Geary, S., Tamura, M. and Kaspar, R. (1998) Nucleic Acids Res., 26, 2565- 2571.
- Special thanks to Masayuki Nashimoto for his extensive work in developing the heptamerguided cleavage method, and for his help on this project.