Benjamin E. Farnsworth and Dr. Gregory F. Burton, Chemistry and Biochemistry
The human immunodeficiency virus (HIV) targets immune cells, specifically CD4 T Cells. Infection of these cells by the virus eventually renders the immune system incapable of proper function, and ultimately results in the death of the patient. Highly active anti-retroviral drug therapies (HAART) have been developed that interfere with the virus replication machinery. HAART treatment produces a dramatic drop of virus particles detectable in the blood. However, HAART has not been able to eradicate the virus. Long-term reservoirs of infectious virus have been discovered in CD4+ T cells, macrophages, central nervous system microglial cells, seminal fluid cells, and thymocytes (1). Other immune cells are potential reservoirs.
It has been suggested that follicular dendritic cells (FDC) have potential to be HIV reservoirs (2). FDC are highly specialized immune cells found in the germinal centers of most secondary lymphoid tissue. Here in the lymphoid tissues, FDC trap and retain antigen (Ag) on the surfaces of their dendritic processes. Trapping of Ag by the FDC creates a repository of protein that aids in long term humoral immune memory. In HIV infection FDC trap and preserve intact virus which retains its ability to infect T Cells. It is for this reason the FDC has been implicated as a long term reservoir for HIV infection (3).
The complex nature of Ag trapping by FDC is not well understood. Human tissue was not available for experimentation, so I purposed to use a murine (mouse) model to mimic human lymphoid tissue and explore the ratio of different viral sub-species on the FDC. After I submitted my proposal, lymph node biopsy tissue from an HIV infected patient was kindly provided by Dr. Timothy Shacker and Dr. Ashely Haase from the University of Minnesota. This presented a better format for addressing the aims of my proposal.
The method employed to understand how the FDC traps virus was to examine the variety of virus recovered from the FDC. DNA sequence analysis was used to compare relationships of individual virus and compile an evolutionary tree. These data form a snapshot of what is happening in the body during HIV infection. To compare genetic changes in virus trapped on FDC, I sequenced a 711 base pair region of the HIV pol gene which codes for the viral reverse transcriptase (RT) enzyme.
Materials and Methods
Biopsy tissue was digested with proteolytic enzymes and FDC were isolated. Viral RNA was converted to DNA by RT-PCR. Nested-set PCR was used to select and amplify viral RT cDNA. Amplification products were ligated into the plasmid vector PCR3.1 (Invitrogen), and these plasmids transformed into STBL2 bacteria (Invitrogen) by heat shock. These bacteria were gown to amplify the RT gene. Sixteen individual clones were selected, plasmids isolated using DNA Mini-prep (Qiagen), and DNA sequenced by Big-Dye termination (Applied Biosystems).
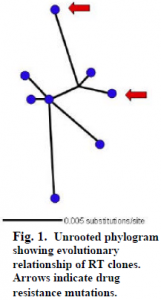
SEQUENCER (Gene Codes Corp.) was used to edit sequences. Nucleotide alignment was done on 11 clones in CLUSTAL W (IGBMC), and edited with in MacClade 4 (Sinauer Associates, Inc.). The best-fit evolutionary model selected in MODELTEST was TrN. A heuristic search in PAUP* (PPC/Altivec) was then used to construct an unrooted phylogram (Fig. 1).
Results and Discussion
Eleven sequences were obtained and analyzed. Of these, three had at least one random nucleotide deletion causing a shifted reading frame. The eight remaining sequences contained a large open reading frame consistent with the HIV RT gene. Analysis of these sequences yielded two clones with possible drug resistance mutations. These were S68N and M184T (the latter is resistant to 3TC, lamivudine). An unrooted phylogram of these eight clones suggests an evolutionary link, through a common ancestor, between these two mutations (Fig. 1, arrows).
These data provide an exciting base to more fully understand the FDC and its role during an HIV infection and concomitant drug therapies. For example, sequencing DNA obtained from T cells recovered from the same biopsy sample will show the diversity that exists between virus from infected T cells and infectious virus trapped on the FDC. Patient drug treatment history can be used to establish a time line. This subject received treatment for six months. If the subject was treated with lamivudine the date that this treatment was initiated or terminated will reveal how long the virus has been located on the FDC. Another way to understand what is happening on the FDC is to examine additional samples. If tissue from the same patient can be collected from a new biopsy of the same site, HIV sequence data can be obtained and a thorough comparison of the lymphatic microenvironment over time can be conducted.
This study provides data that shed further light on the role of FDC and support their function as an HIV reservoir. Through understanding viral reservoirs, treatments can be developed that will prolong the life of those suffering from HIV infections.
References
- T. W. Chun, et al., Nature. 387,183 (1997).
- G. F. Burton, et al., Sem. Immuno. 14, 275 (2002).
- B. A. Smith, et. al., J. Immuno. 166, 690 (2001).
