Gregory C. Chipman and Dr. Daniel L. Simmons, Chemistry and Biochemistry
My efforts were concentrated in assisting Phillip M. Robertson, a graduate student in the Department of Chemistry and Biochemistry, in researching the cellular action of a family of drugs, nonsteroidal anti-inflammatory drugs. Nonsteroidal anti-inflammatory drugs (NSAIDS) include aspirin, ibuprofen, diclofenac, and other pharmacologically important agents. They have many applications, including reduction of inflammation and pain. These drugs have also been shown to induce apoptosis, or programmed cell death, in tumor cells. It has been generally accepted that NSAIDs function by inhibiting prostaglandin synthesis by enzymes called cyclooxygenases (COX). However, studies have questioned whether all NSAID actions are exerted through the two described forms of cyclooxygenase COX- I and COX-2. For instance, mice in which the COX-2 gene was disrupted showed unaltered inflammation response.1
To determine the cellular product involved in NSAID induced apoptosis (NIA), chicken embryo fibroblasts (CEF) containing a temperature sensitive mutant of the rous sarcoma virus (RSV) were treated with diclofenac (100-125 pM) and simultaneously treated with potential mediators of the NIA pathway. RSV-CEF cells are normal at 42(C but transform into cancer cells at 35(C, which makes the cell line a good model for studies on cancer cells. This approach was designed to identify possible substrates and/or products of the principle enzyme involved in NIA. If the agent being tested was a natural substrate of the enzyme, it would bind in competition with the NSAID and rescue the cells from apoptosis. If it were a product of the enzyme, it would rescue the cell from apoptosis despite inhibition of its production by the presence of the NSAID). Potential agents tested included Prostaglandins, the natural products of cyclooxygenase activity, and fatty acids, the natural substrates.
Apoptosis was characterized by observing the morphology of the cells and quantified by the MTT assay, which measures mitochondrial viability. MTT is administered and viable cells metabolize the MTT to a purple color. Coloration is measured using a spectrophotometer. Of all the agents tested, linolenic acid, adrenic acid, and oleic acid were possible substrates. All of these long chain fatty acids rescued cells from apoptosis up to certain concentrations and promoted apoptosis at greater concentrations. Interestingly, the apoptosis was evident by morphology of the cells but not by the MTT assay, suggesting a different time course of expression of morphological changes and disruption of mitochondrial viability.
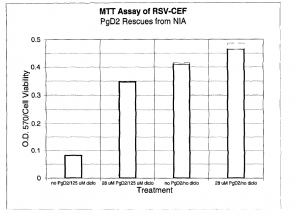
Of the possible products tested, prostaglandin D2 showed clear signs of rescue in both morphology and MTT assays. (Figure 1) Cells were treated with 125 μM diclofenac and 28 μM PgD2. This suggests that inhibition of PgD2 synthesis is involved in the NIA pathway.2
References
- S.G. Morham et al. (1995) Prostaglandin Synthase 2 gene disruption causes severe renal pathology in the mouse. Cell 83, 473-482.
- This work was done with Phillip Robertson, a graduate student in the Department of
Chemistry and Biochemistry. - Figure 1. PgD2 rescues from apoptosis. Metabolized MTT was measured in a spectrophotometer
at 570 nm.