Matthew Burnham and Dr. Brad Bundy, Department of Chemical Engineering
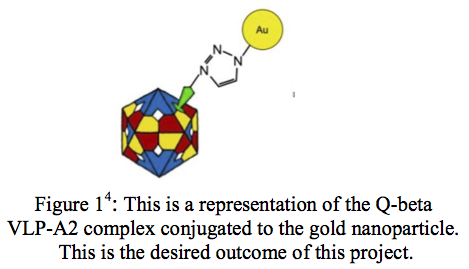
A Virus-like Particle (VLP) is a nanostructure composed of many coat proteins. They differ from real viruses in that they are synthesized to be non-infectious. VLPs have many useful applications in vaccines, drug delivery, gene therapy, and material science1. Last year, I attempted to create a high resolution image of the VLP Q-beta. This was to be accomplished by making modifications to the virus’ structure after its creation. Conjugating a gold nanoparticle to the Q-beta VLP-A2 protein complex would, in theory, provide a site of orientation and allow us to image the VLP (Figure 1). Although the goal of obtaining an accurate image was not reached, steps were made towards one day reaching it.
I worked on this project with Mark Smith, a graduate student in Chemical Engineering. We began our experimentation by obtaining the self assembling coat proteins via a process called cell-free protein synthesis (CFPS)1. Once we had our VLP intact, we attached to its surface a modified A2 protein2. This protein was modified through the incorporation of the unnatural amino acid (UAA) p-propargyloxyphenylalanine3. This is important because it is this UAA that is the potential site of attachment of the gold nanoparticle.
Before we began experimenting with the gold nanoparticle, because gold is really expensive, we decided to first see if the UAA incorporated into the A2 protein was available for attachment. We decided to do this because of the nature of protein structures. After proteins are coded, they ‘fold’ into their final structure. We therefore needed to verify that the UAA was located on the outer surface of the protein and not within the structure. To do this, we reacted the Q-beta A2- VLP complex with a polyethylene glycol (PEG) derivative. We thought this molecule to be the perfect candidate to check for the A2 availability because it has the same nitrogen group as the gold nanoparticles and is relatively cheap.
After reacting the VLP complex with PEG, we ran a protein gel, which separates proteins based on size. If the gel showed two resulting bands instead of one, then we would have been fairly confident that the complex did indeed conjugate with PEG. Once the gel was finished, we performed an autoradiogram, which is sensitive to the radiation we added to the protein and created an image of the protein gel separation. An autoradiogram, however, takes four to six weeks to develop. Due to our confidence in the success of this experiment, we thought that this would be an effective method to verify the conjugation of the VLP and PEG, because an autoradiogram can detect low levels of radiation and could therefore detect attachment a low efficiencies. After developing our autoradiogram, the only thing we discovered was that our experiment did not work.
This was a difficult time in the experimental process, because we were unsure whether there was something wrong with our experimental methods or something wrong with our experimental design. At this point, due to the lengthy nature of the experimental process, both with the preparation of the VLP and the autoradiogram, we were not willing to attempt another autoradiogram and therefore had to use a different method to verify the VLP-PEG conjugation.
To speed up the process, we decided to change our experimental design by testing the accessibility of the UAA on the A2 protein without the entire VLP complex. This decreased the amount of time we had to spend growing proteins and allowed us to focus our efforts on the task at hand. We discovered from our autoradiogram that PEG was not a convenient molecule to use in this experiment because it greatly interfered with the ability of the proteins to separate within the protein gel. We therefore began to test the attachment with fluorescent fluorophores. The fluorophores would allow us to verify conjugation by measuring the fluorescence of the molecules compared to a control. We would also compare this to the amount of radiation given off by the molecules, which would allow us to see yield amount and concentration of the molecules in solution.
Our first attempt at conjugating the A2 protein with fluorophores yielded promising results. Our data showed that two of the seven A2 mutants that we were working with showed some conjugation. Frustration came, however, when we attempted to replicate our results. After three attempts at verifying conjugation, which were unsuccessful, we had to conclude that that our data was inconclusive. Despite this disappointing result, there is still reason to believe that the UAA on the A2 protein is accessible for attachment. Many methods exist to verify conjugation with the A2 protein and efforts are still being made to conjugate the gold nanoparticle with the VLP-A2 protein complex.
It has been a very rewarding experience participating in this research. I have learned a great deal about the scientific method and have been able to increase my technical skills and problem solving ability. I have learned how to handle disappointment and how to look past disappointment to look for solutions.
References
- Bundy, Bradley C., Marc J. Franciszkowicz, and James R. Swartz. “Escherichia Coli-Based Cell-Free Synthesis of Virus-Like Particles.” Biotechnology and bioengineering 100.1 (2008): 28-37.
- Smith MT, Varner CT, Bush D, Bundy BC. “The Incorporation of the A2 Protein to Produce Novel Q[beta] Virus-like Particles Using Cell-free Protein Synthesis.” Submitted and accepted, pending publication: Biotechnology Progress. Oct 2011.
- Bundy, B. C.; Swartz, J. R., “Site-Specific Incorporation of p-Propargyloxyphenylalanine in a Cell-Free Environment for Direct Protein-Protein Click Conjugation.” Bioconjugate Chemistry 2010, 21, (2), 255-263.
- All images modified from a graphic by Derek Bush. Used with permission.