Erik J. Petersen and Dr. Terry S. Elton, Chemistry and Biochemistry
A comparison of several published hAT1R cDNA sequences revealed that, although these clones shared an identical open reading frame, they differed in portions of their 5′-UTRs.1 Therefore these data suggest that alternative splicing events combine various 5′-UTR exons with the exon that harbors the hAT I R coding region.
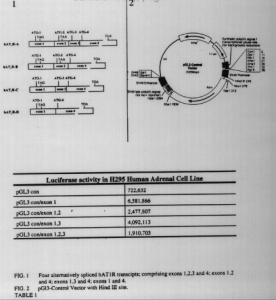
Experiments utilizing the 5′-RACE technique indicated that at least four distinct alternatively spliced hAT1R mRNA transcripts are transcribed in human lung tissue (e.g.. hAT I R transcripts can be comprised of exons 1,2,3 and 4; exons 1,2 and 4; exons 1,3 and 4; or exons 1 and 4) (Fig. 1) all of the alternatively spliced hAT1R transcript contain exons 1 and 4 (i.e., the entire coding region). Thus, although alternative splicing events combine various 5′-UTRs, all four transcripts are thought to encode for identical hAT1R proteins since the coding region is harbored on exon 4 (Fig. 1). These results also suggest that, at least in human lung, there is only one functional promoter in the hAT1R gene since all of the hAT1R transcripts characterized contain the same first exon.
To investigate the translational activity of the alternatively spliced exons the various spliced exons (i.e. exons 1,2 and 3. 1, 2; 1, 3-, and exon 1) were subcloned into the Hind IH site of the pGI3-control vector. (Fig.2) This vector harbors an SV40 promoter driving luciferase cDNA. The data collected from these experiments are recorded in table 1. Exon 1 increase translation efficiency 8 times, where as exon 2 decreases translational efficiency of luciferase CDNA. When reading the sequence of exon 2 an ATG start and TAG stop codons our found. We hypothesize that the ATG start codon allows ribosomes to transcribe an eight amino acid peptide until it reaches the TAG stop codon. Thus once the ribosome reaches the stop codon it falls off the DNA,hindering transiation. Consequently, hATIR production can be manipulated; by increasing or decreasing the translation efficiencies of the hAT1R exons.
References
- Baogen Su, 1994. Biochemical and Biophysical Research Communications 204: 1039-1046.