Jacob Venesky and Dr. Greg F. Burton, Department of Chemistry and Biochemistry
My ORCA research project was designed to assess the level of diversity found in a patient’s body. When a doctor prescribes drugs to combat HIV, he or she must first assess whether the patient houses drug-resistant strains. This is usually done by extracting blood from the patient and sampling the HIV found within. There is a general assumption made that if there are no drugresistant strains of HIV found in the blood, then the prescribed drugs will be effective at controlling the virus. However, if the HIV found in the blood of the patient was not fully representative of the level of HIV diversity found throughout the patient’s entire body, the drug treatment may not be effective. In order to answer that question, samples of HIV extracted from a different part of the body would need to be sampled and compared with HIV samples from the blood. My project specifically revolved around determining the diversity of HIV found within a sample of cells known as follicular dendritic cells. Follicular dendritic cells, or FDCs, are of especial interest to HIV research in that they are a reservoir in the body for HIV. By sampling and classifying the HIV quasi-species that are found on FDCs, one can then compare them to virus quasi-species found circulating in the blood. The benefit of this comparison would be to clarify whether a resistance screening only sampling HIV from a patient’s blood would suffice in determining drug resistances. My task in this question was to determine the DNA sequences of the HIV found on FDCs from one patient so that those sequences could later be compared with samples of HIV found in that same patient’s blood.
The process of extracting and sequencing HIV DNA requires patience and consistent effort. There are about ten different steps to bring a sequence of DNA from the cell where it resides to the computer screen. Because of my unfamiliarity with the techniques needed to sequence a sample of HIV, significant time was spent mastering the procedures needed to accurately obtain the data.
Problems began at the outset. The first month of my learning was spent repeating the initial step, which is known as amplification. In order to take a sample of just ten individual HIV from the FDCs, you must amplify the small sample and separate the individual clones from one another. The process is no small task. It is similar to searching for ten needles in a haystack, and then trying to distinguish the differences between each needle. The only problem is that you can’t see the needles nor the haystack. Each protocol must be done to perfection in order to see the desired results. All of the techniques were new to me to begin with, but after months of repeating the same procedures, I became an expert at each task. I constantly had to collaborate my failed findings with my mentor, Dr. Burton, to try and discern what might have gone wrong. The entire process taught me to think more critically about what I was doing. It required me to do more than follow a set of instructions. Dr. Burton helped in this process by not giving me the obvious answers right away. I had to explain to him what I felt went wrong or what the possible sources of error might be. More often than not, I ended up with the answer without Dr. Burton having to say a word. Although I had more setbacks in my project than I would have liked, I came out of those setbacks not only being able to overcome them, but also with the ability to look for new paths around future barriers, whether in research or elsewhere in my life.
After months of trial and error, the I was finally able to amplify and separate the different clones I had made of the HIV found on the patient’s sample of follicular dendritic cells. The last step I needed to perform manually in the lab was to prepare the ten samples I had cloned for sequencing in the BYU Sequencing Center. As fortune would have it, I was able to obtain highly accurate sequences from my ten samples on the first try.
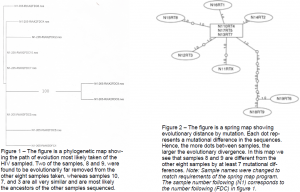
After sequencing my ten samples, my lab skills were left aside to focus on manipulating the newly gathered data through the use of various computer programs. I first had to trim and align each sequence, a process similar to lining my ducks in a row to be observed for differences. I came to Dr. Burton with these aligned sequences, and together we inserted the data into a program that created a phylogenetic tree, or a picture showing the evolutionary relationship between the ten HIV regions sequenced (see figure 1). The data was also demonstrated in a springer map, which shows each mutational difference represented as a dot (see figure 2).
Although my samples represented just one region of the patient’s body, the data obtained are a necessary component needed to answer the question: are current methods of testing HIV patients (sampling HIV from the blood) adequate at assessing true drug resistance? With this data in hand, further work can now be done on sampling HIV from the same patient’s blood. With HIV sequences from these two sources, the data can then be compared to determine if there are differences strong enough to merit a change in the way doctors assess their patients.