Emily Parker Updegraff and Dr. Merritt B. Andrus, Chemistry and Biochemistry
Chemotherapy is one of the primary methods of combating cancer, and if it fails, the patient may be left with few options for treatment. Multi-drug resistance (MDR) is a clinical condition manifested by the failure of chemotherapy. It is primarily due to the expression of Pglycoprotein (Pgp), a membrane-bound small molecule transport protein which is found normally in some cells but is greatly over-expressed in drug resistant cancer cells1. After chemotherapy drugs diffuse into the cell, Pgp effluxes them out against their concentration gradient, thus imparting drug resistance. The general belief is that the most direct and practical way of reversing MDR is through developing Pgp inhibitors. To accomplish this, we must learn more about the structure and mechanism of transport of Pgp.
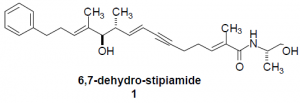
The basic structure of Pgp is a pore composed of twelve transmembrane helices. The protein contains binding sites at helices six and twelve, but their exact locations within the protein are unknown. A definitive model of Pgp’s three-dimensional structure has been hard to obtain because direct structural analysis of membrane-bound proteins is difficult. Determining the structure of Pgp, then, must be done by indirect experimental techniques2. My research has involved synthesizing compounds which are used to explore the structure of Pgp. MDR reversal agent 1, developed by Andrus et al3, is known to bind Pgp. I have synthesized homodimers consisting of two molecules of compound 1 linked together by a polyethylene glycol tether of variable length (Compound 6). By measuring the MDR reversal potency in cancer cells treated with these homodimers, the approximate distance between TM 6 and TM 12 can be discovered. The dimer with highest potency will indicate that the tether length used is the approximate distance between the two binding sites. This technique will establish whether the helices are adjacent or opposite each other in the pore and will provide the first map of the binding sites’ relative locations in the structure of Pgp.
The following scheme shows the synthetic route to the homodimers. We synthesized dimers of tether lengths n=0, 2, 5, 8, and 12. Diamines of lengths n=0 and 2 were commercially available, but the longer diamines had to be synthesized from polyethylene glycols. The n=5 glycol was commercially available, but the n=8 and 12 were not, and their synthesis presented the greatest obstacle in this research.
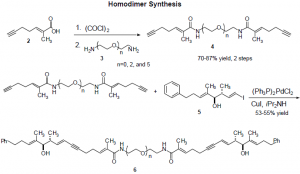
Several leaving groups, including chloride, bromide, and tosylate, were tried as leaving groups in the Williamson ether synthesis to yield the n=8 and 12 glycols. These gave very low yields, up to only 14%. We then used a mesylate leaving group, sodium hydride as the base, and heated the reaction mixture to reflux to obtained the desired glycols in 73% yield for the n=8 and 59% for the n=12. We used a synthetic route similar to that of Jenneskens4, but our conditions for are unique in our use of a mesylate leaving group and in heating the reaction mixture to reflux.5
All five homodimers have now been synthesized and sent to our collaborators at NIH and Purdue University for biological assays; we are currently awaiting those results. Data from the biological assays will establish the distance between Pgp’s binding sites, giving us a better picture of the protein’s three dimensional structure. This knowledge will be important because it could aid in discovering effective treatments for drug resistant forms of cancer.
References
- Ferry, D.R.; Traunecker, G.; Kerr, D.J. Eur. J. Cancer 1996, 32A, 912-920.
- Higgins, C. H.; Callaghan, R.; Linton, K. J. Cancer Biology 1997, 135-142.
- Andrus, M.B.; Leponre, S.D.; Turner, T.M. J. Am. Chem. Soc. 1997, 12159-12169
- Jenneskens, L.W.; Keegstra, E. M. D.; Zwikker, J. W.; Roest, M. R. J. Org. Chem. 1992, 6678-6680.
- Many thanks to Timothy Turner for his help with the synthesis of the longer polyethylene glycols.
