Brandon Carter, Dr. David B. Dahl, Department of Statistics
Introduction
Cluster analysis is an important exploratory data analysis technique used in a wide variety of fields. Cluster analysis seeks to discover a natural grouping of the data, where items in the same cluster or group are more similar than items from different clusters. Through our research, we developed a novel method for cluster analysis which takes pairwise distance information as input. Our new method improves upon traditional cluster analysis methods which also take pairwise distance information as input, such as hierarchical clustering. Our method, cluster analysis via random partition distributions (CaviarPD) is based on probability distributions and therefore allows the user to discuss probabilities that items are clustered together. Hierarchical clustering, a heuristic method, does not allow for any probability statements about how items are clustered. CaviarPD also gives a better visual of the estimated clustering. Furthermore, through case studies where the true groupings of the data are known, CaviarPD performs better at estimating the true grouping of the data as compared to hierarchical clustering. We now explain hierarchical clustering and how our new method CaviarPD compares to this traditional method.
Methodology
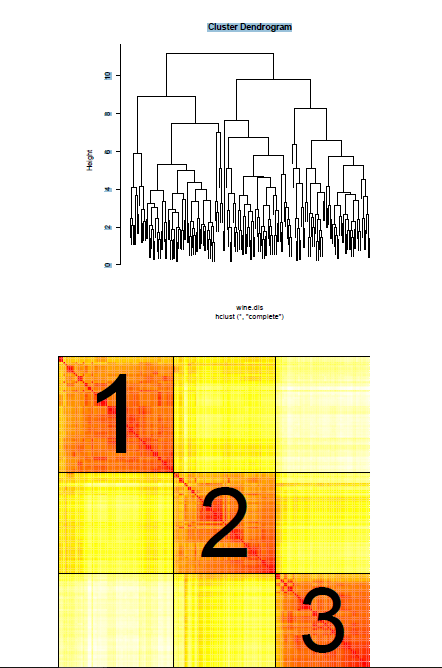
Hierarchical clustering is a heuristic algorithm that begins with each item in its own cluster and successively merges the most similar clusters until all items are in a single cluster. The criteria for defining cluster similarity is called linkage and clusters that are the most similar are merged first. The succession of merges results in a hierarchy of clusters as shown in Figure 1, a clustering tree of a dataset containing the chemical makeups of wines from 3 different cultivars. In order to produce an estimate of the partition of the dataset the tree is cut at a specific height which creates distinct clusters of the items in the dataset. There is no consensus to choosing the height that a clustering tree should be cut. Statistical textbooks even admit that choosing where to cut the tree is often done subjectively by examining the structure of the tree.
Our development of CaviarPD is motivated by the Ewens-Pitman Attraction (EPA) distribution, a probability distribution over partitions. A constructive definition of the distribution is that observations are sequentially allocated to subsets of a partition with probability proportional to the attraction of the item to that subset. The probability of an item being allocated to a new subset is governing by the mass parameter. The higher the mass parameter the more likely an item will be allocated to a new subset. Draws are obtained from the EPA distribution and are summarized by an expected pairwise allocation matrix (EPAM). Though our method is not Bayesian, estimation techniques commonly used in Bayesian nonparametrics are employed in order to estimate the partition of the data which best represents the EPAM.
A plot the EPAM of the wine dataset is given in Figure 2. The resulting partition estimate is highly dependent on the mass parameter in the EPA distribution. We developed and algorithm to select the mass parameter that best separates the items into distinct groups. Thus, unlike hierarchical clustering, CaviarPD has a built in method to select the number of clusters in the partition estimate.
Results
We used the adjusted rand index (ARI), a measure of similarity between partitions on a scale of 0-1, to compare how well the two methods estimated the true partitioning of the data. Because there is no default method to cut the clustering tree, we selected the cut of the tree that maximized the ARI. For the wine dataset, the very best cut of the tree produced a grouping of the data with 7 clusters and an ARI of .66. CaviarPD, using our mass selection algorithm produced a clustering estimate with 3 clusters and ARI of .79. Thus, CaviarPD both estimated the correct number of clusters and over all produced a better estimate of the true clustering of the wine dataset.
Discussion
CaviarPD in nearly all of our case studies performed better than the very best possible cut of the tree from hierarchical clustering. CaviarPD has a built in method for selecting the mass parameter, therefore is not as subjective as tree cutting in hierarchical clustering. Furthermore, CaviarPD allows the user to discuss probabilities that items are clusters together for a given value of the mass parameter.
Conclusion
CaviarPD not only performs better than hierarchical clustering in estimating the true partition, but also gives a better mathematical and visual representation of clustering results.
Figure 1 – A clustering tree of the wine dataset. The leaves of the tree represent individual items in the hierarchy of clusters. Items that are more similar are merged first, i.e. at a lower height of the tree. Similarity between clusters is relative to the vertical height of merged branches.
Figure 2 – A confidence plot of the wine dataset. The color of each cell represents the probability that two items are in the same cluster. The estimated clusters are delineated and labeled. 0 2 4 6 8 10 Cluster Dendrogram