Millicent Campbell, David Michaelis, Department of Chemistry and Biochemistry
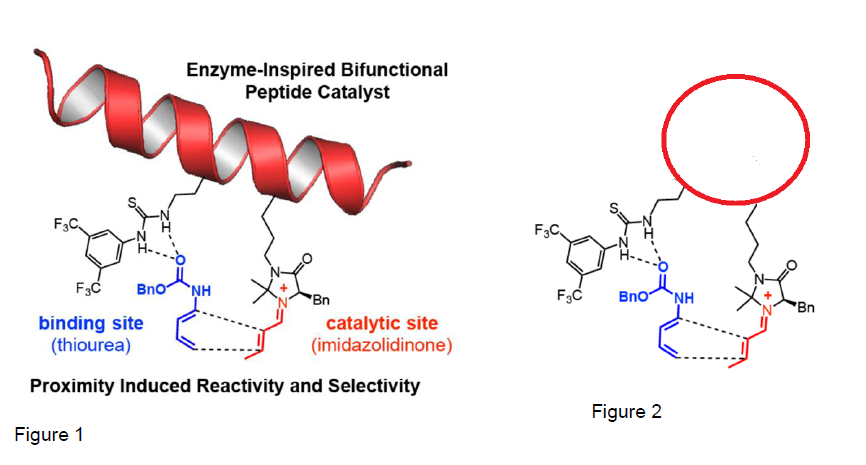
Enzymes found in nature are more efficient catalysts than those used in organic chemistry labs. However, natural enzymes are not ideal for organic synthesis because they only make one product and only work in specific conditions. The Michaelis lab designed a catalyst capable of mimicking enzyme-like reactivity in the lab. This catalyst consists of an alpha helix with an imidazolidinone catalyst and a thiourea catalyst (see figure 1). The imidazolidinone catalyst and thiourea catalysts are attached adjacent to each other on the alpha helix, which acts as a rigid scaffold that can mimic the proximity effects seen in natural enzymes. In the past, we have shown that this alpha helical catalyst can catalyze a Diels-Alder reaction, which is important to pharmaceutical research, faster than other catalysts. In the past few months, we have also attached a TEMPO catalyst to the alpha-helical scaffold. This catalyst is currently used only in academia because it is too expensive to use on large scales needed in industry. Attaching it to our alpha helical scaffold would increase the recyclability of the TEMPO catalyst and reduce its cost.
Our Catalyst works because it keeps the two organic catalysts stable via a scaffold. My project focused on the idea that holding the two peptides close together would induce proximity effects to catalyze a reaction. I proposed, that by attaching the two ends of the peptide that forms a helix, together to form a cyclic structure (see figure 2). This would create a rigid structure capable of inducing similar proximity effects. I had planned to find a way to design the peptide, to synthesize the peptide, and attach two organic catalysts to the cyclic scaffold.
I began by designing the sequence of the cyclic structure with amino acids that have a propensity towards forming a beta turn. The peptide consisted of seven amino acids, starting with aspartate, phenylalanine, lysine, lysine, phenylalanine, glycine, and methionine. I capped the lysine with two different caps, so that I could functionalize them with the imidazolidinone and thiourea catalysts that we had previously attached the Michaelis alpha-helix. I used microwave assisted solid phase peptide synthesis to synthesize the peptide. I checked the integrity of the synthesis via liquid phase mass spectrometry. This also allowed me to see contaminants in the peptide.
After this, I tried attaching the organic catalysts by deprotecting the individual protecting groups on the lysine amides while the peptide was on resin. I also tried cyclizing the peptide without the attached organic catalysts. I was able to attach the organic catalysts before cyclization easily. However, I was unable to cyclize the peptide after this. I was also unable to cyclize the peptide on resin without the catalysts attached.
After trying various methods of attaching the catalysts, I tried various resins. However, I was unable to cyclize the peptide on rink amide or HMBA resins. These resins would have different effects because rink resin attaches to the peptide via an amide bond, while the HMBA attaches via an ester bond. This means that when the peptide is cleaved from the resin, it would end in either an ester or an amide bond. The cyclization of the peptide mechanism would involve the amino terminal attacking the initial amino acid carboxyl terminal. A Hydroxyl group is more stable than an amine leaving group. We believed that building the peptide on the HMBA resin would make the cyclization on resin more effective because it would create a hydroxyl leaving group rather than an amine. Despite this, my cyclization procedure was unsuccessful.
Another thing that contributes to cyclizations is purity of the sample, and peptide concentration. The peptide is far more likely to cyclize at low concentrations in pure solutions. This is because the solvent pushes the peptides into a cyclic form. I attempted cyclizing the peptide at 10 mmol, 50 mmol, and 100 mmol concentrations. I was unable to cyclize the peptide this way either.
After discussing the methods with Dr. Michaelis, he suggested that the current approach did not account for the high energy barrier for cyclization. I am currently working on a project that focuses on using molecular modeling to show the interactions between two catalysts mounted on a peptide scaffold. After we finish developing the programs to analyze the peptide, we will be able to use these same programs on the cyclic peptide. It is important to develop these programs on the original peptide because we have data that we can confirm accuracy of the programs with. These programs are based on using programs such as GAMESS and AMBER tools to perform calculations on the fulton supercomputer.
By researching the cyclic peptide, we learned that the energy barrier for cyclization is high enough that various methods of cyclization are rendered ineffective. I would need to research more to find a way to cyclize the peptide. One way to do this is by using computer modeling programs and the supercomputer.